You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yang, Y.; Huang, J.; Sun, Q.; Wang, J.; Huang, L.; Fu, S.; Qin, S.; Xie, X.; Ge, S.; Li, X.; et al. miRNAs to Cu Toxicity. Encyclopedia. Available online: https://encyclopedia.pub/entry/41394 (accessed on 27 December 2025).
Yang Y, Huang J, Sun Q, Wang J, Huang L, Fu S, et al. miRNAs to Cu Toxicity. Encyclopedia. Available at: https://encyclopedia.pub/entry/41394. Accessed December 27, 2025.
Yang, Ying, Jiu Huang, Qiumin Sun, Jingqi Wang, Lichao Huang, Siyi Fu, Sini Qin, Xiaoting Xie, Sisi Ge, Xiang Li, et al. "miRNAs to Cu Toxicity" Encyclopedia, https://encyclopedia.pub/entry/41394 (accessed December 27, 2025).
Yang, Y., Huang, J., Sun, Q., Wang, J., Huang, L., Fu, S., Qin, S., Xie, X., Ge, S., Li, X., Cheng, Z., Wang, X., Chen, H., Zheng, B., & He, Y. (2023, February 19). miRNAs to Cu Toxicity. In Encyclopedia. https://encyclopedia.pub/entry/41394
Yang, Ying, et al. "miRNAs to Cu Toxicity." Encyclopedia. Web. 19 February, 2023.
Copy Citation
Environmental metal pollution is a common problem threatening sustainable and safe crop production. Heavy metals (HMs) cause toxicity by targeting key molecules and life processes in plant cells. Plants counteract excess metals in the environment by enhancing defense responses, such as metal chelation, isolation to vacuoles, regulating metal intake through transporters, and strengthening antioxidant mechanisms. microRNAs (miRNAs), as a small non-coding RNA, have become the central regulator of a variety of abiotic stresses, including HMs.
heavy metals
miRNA
toxicity response
1. Introduction
The rapid growth of industrialization and the large-scale use of chemical fertilizers and pesticides have led to the continuous increase in heavy metal content in the soil. In plants, copper (Cu), zinc (Zn), iron (Fe), and manganese (Mn) are trace elements necessary for plant development and growth, but excessive accumulation can cause cell damage [1]. Some other metals such as cadmium (Cd), chromium (Cr), lead (Pb), aluminum (Al), arsenic (As), and mercury (Hg), as non-essential elements, are toxic even at low concentrations [2]. Studies have reported that heavy metal stress can inhibit activity of antioxidant enzyme systems and differential expression of a large number of proteins in plants, weakening photosynthesis, accompanied by a series of phenotypes such as suppressed root development and leaf senescence or even necrosis [3]. In order to avoid the destructive consequences of heavy metal toxicity, plants have developed corresponding coping mechanisms to resist heavy metal stress [3]. Plants can reduce heavy metal concentrations in their bodies by limiting the uptake of heavy metal ions and stimulating metal efflux, as well as by complexation of metal ligands such as glutathione (GSH), metallothionein (MT), and phytochelatins (PCs) [4][5][6]. In addition, the antioxidant defense mechanism will also be activated to reduce elevated reactive oxygen species (ROS) levels, thereby reducing oxidative damage [7][8]. A large number of studies have shown that gene expression plays a very important role in regulating the tolerance of HMs or individually regulating various stress response genes to form a gene network. Some functional group genes encode metabolites such as amines, alcohols, and sugars, which also play a crucial role in heavy metal stress tolerance [9][10].
With the discovery of small RNAs, there is growing interest in the importance of post-transcriptional gene regulation of microRNAs (miRNAs) in plant development and responses to environmental stresses. miRNAs are an extensive class of small noncoding (19~24 nt) RNAs molecules [11]. In plants, mature miRNAs are produced through a multistep process including the transcription, precursor processing, methylation, and assembly of miRNA-induced silencing complex (miRISC) [12]. Then, the mature miRNAs lead the RISC to target the complementary mRNAs, which play a vital post-transcriptional regulatory role in gene expression by target mRNA cleavage or translational inhibition [13]. In recent years, a crescendo of miRNA studies has demonstrated that miRNAs play important roles in tissue development and differentiation, phytohormones signaling, secondary metabolite production, and biotic and abiotic stress [14][15]. miRNAs affect multiple processes of plant growth, development, and response to stress by up-regulating or down-regulating their expression [16][17]. miR165/166 and miR394 have been shown to be involved in shoot apical meristem (SAM) development, including the direct post-transcriptional regulation of key SAM-related genes, which in turn maintains SAM development [18][19]. In addition, studies have shown that miR319 has a conserved regulatory function in leaf development. For example, ectopic upregulation of miR319 resulted in dramatic changes in tomato leaf size and shape [20]. CUC1 and CUC2 mRNAs accumulate in the axil of leaf primordia and play a key role in the establishment of axillary bud meristems. The regulatory mechanism of miR164-CUC1/CUC2 may be related to LAS-mediated initiation of axillary bud meristems [21]. In addition, miRNAs play important roles in plant root development. miR160 acts as a key controller and cleaves ARF10 and ARF16 transcripts during plant root cap formation [22]. In contrast, miR167 plays an active role in adventitious root formation, while miR156 and miR172 are well-studied miRNAs involved in floral control [23][24]. Overexpression of miR172 can promote flowering time in both monocotyledonous and dicotyledonous plants. In contrast, the expression level of miR156 decreased gradually from sowing to flowering, while upregulation of miR156 resulted in delayed flowering transition. These findings suggest that miRNAs play regulatory roles in different developmental transitions by mediating specific signaling pathways. In different models and crop plants (such as Arabidopsis, wheat, rice, maize, and barley), miRNAs regulate gene expression during different stress responses (drought, heat, salinity, cold, nutrition, and pathogens). In addition to plant growth and development, the role of conserved miRNA target modules is also critical for conferring stress tolerance through integration into metabolic pathways [25]. Studies have confirmed that miR160-ARF, miR156-SPL, miR159-MYB33, miR164-NAC, miR172-AP2, miR394-LCR, miR396-GRF, and miR398-CSD modules play important regulatory roles in different stress environments to mitigate the effects of adverse reactions [12]. For example, in Arabidopsis, increased expression of miR398 enhances plant heat tolerance by negatively regulating the expression of its targets CSD1, CSD2, and the copper chaperone (CCD) of CSD [26]. The highly conserved miR394-LCR module is involved in plant responses to cold stress [27]. The expression of auxin-responsive factors ARF10, ARF16, and ARF17, mediated by miR160 and miR167, resulted in enhanced salinity tolerance of cotton under high salt stress. Overexpression of osa-miR319a exhibited higher tolerance to drought and salt stress by regulating the TCP transcription factor [28].
Increasing evidence has also revealed that miRNA-mediated gene regulation plays a significant role in heavy metal regulatory networks. In addition, high-throughput genome-wide expression profiling has greatly improved the current understanding of the key involvement of miRNA in the toxic response of plant HMs and its targets. A substantial number of heavy-metal-responsive miRNAs have been identified in the Oryza sativa L., Zea mays L., Medicago truncatula L., Hordeum vulgare L., Vitis vinifera L., Brassica juncea L., and many other plants (Table 1) [6][29][30]. In different plant species, many miRNAs have significant differential expression of different HM such as Cd, Hg, Al, As, and Cr. Through the analysis of metal-regulated miRNA target genes, it has also been identified that many miRNAs are involved in the response of plants to HM. The target genes for various processes, including metal absorption and transport, sulfate distribution and assimilation, protein folding, antioxidant systems, and plant hormone signal transduction processes [11]. Researchers searched the Web of Science core collection database for articles related to miRNA and heavy metals such as Cu, Cd, Hg, Al, As, and Cr in the past 10 years, and a total of 568 papers were retrieved. The research fields are mainly distributed in three aspects: Life Sciences Biomedicine, Science Technology, and Physical Sciences. Then, bibliometric analysis was performed using VOS software [31], and four clusters were formed with miRNAs, stress, cadmium, and strategy as the research centers (Figure 1). These keywords are categorized by publication year to deepen the analysis. The VOSviewer software represents the year with the highest number of publications between 2016 and 2019. However, articles from 2011 to 2022 were included in this analysis. The blue circles represent topics that have been intensively studied in the past decade, such as stress response, miRNA, target gene, etc. The results of the two co-occurrence maps showed that miRNAs and stress were most closely related, suggesting that the involvement of miRNAs in stress response is the current research hotspot.
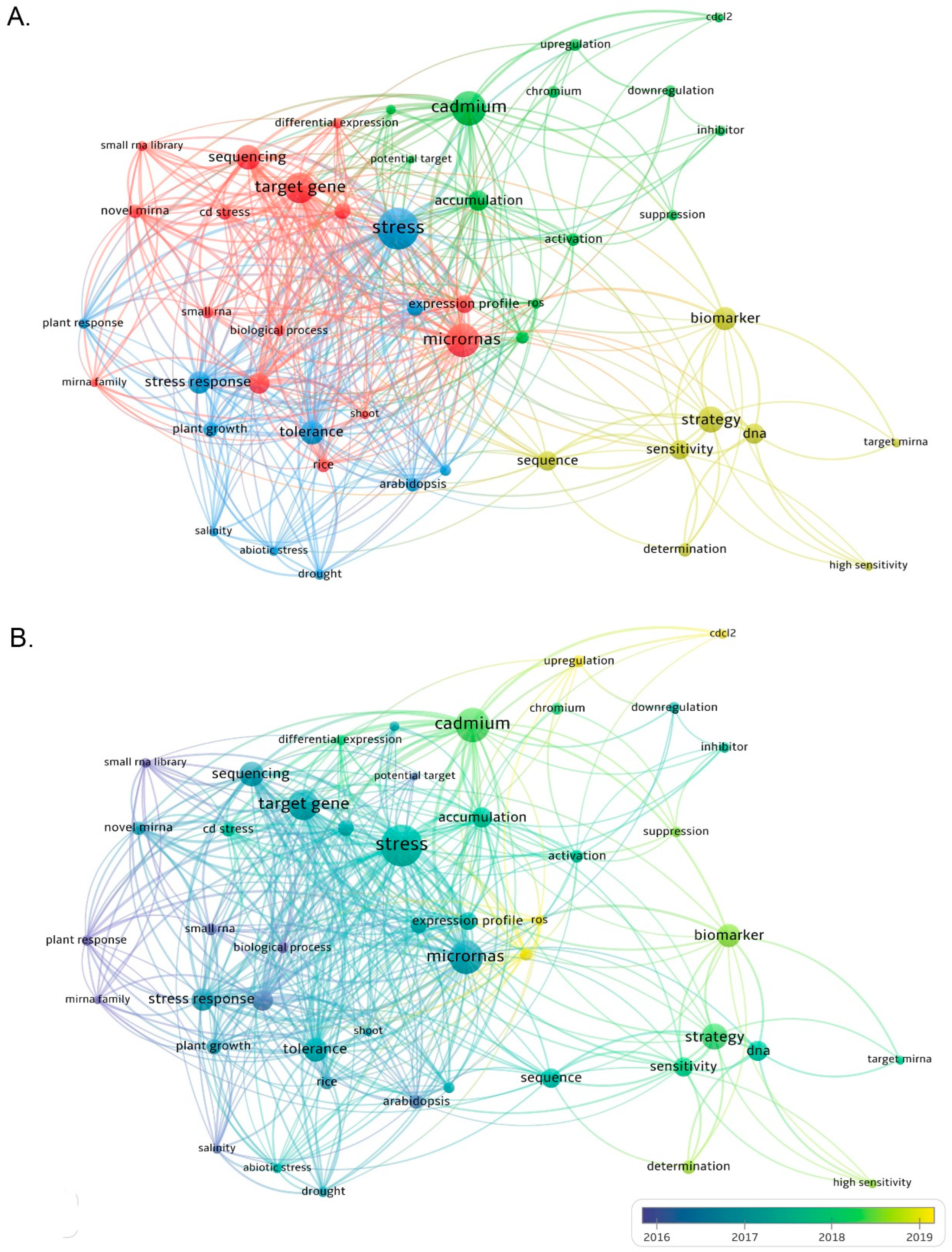
Figure 1. Keyword co-occurrence graph analysis using VOSviewer software. (A) A total of 568 relevant articles were searched from academic papers over the past 10 years using the keywords miRNA, Cu, Cd, Hg, Al, As and Cr, four research centers focusing on miRNA, stress, cadmium, and strategy were formed. (B) Different colors correspond to the year in which the keywords appeared on average, and keywords with blue color presented earlier than those with yellow.
Table 1. List of miRNAs along with their target function during heavy metal toxicity tolerance.
| miRNA | Plant Species | Stress | Target Genes | Target Function | Reference |
|---|---|---|---|---|---|
| miR156 | A. thaliana, V. Vinifera, O. sativa, Nicotiana tabacum L., Miscanthus sinensis A. |
Cu, Cd, Cr, Al | SPL7 | Regulate Cu homeostasis, decrease endogenous ROS | [32][33][34][35][36][37] |
| miR159 | Brassica napus L., O. sativa, N. tabacum, M. truncatula, Saccharum spp | Cd, As, Cr, Al | ABC transporter protein, OPT1 | Regulate metal transporters | [35][36][38][39][40][41] |
| miR160 | A. thaliana, O. sativa, Saccharum spp | Cr, Al | ARF | Regulate auxin signal | [41][42] |
| miR164 | A. thaliana, O. sativa, M. sinensis, Saccharum spp |
As, Cr, Al, Hg | NAC, CUP trancription factors | Signaling pathway, root development, response to oxidative stress | [41][43][44] |
| miR166 | O. sativa, N. tabacum | Cd, Cr | Homeodomain containing protein 4 | Reduced Cd translocation from roots to shoots and Cd accumulation in seeds, decrease Cd-induced oxidative stress | [35][36][45] |
| miR167 | O. sativa, B. juncea, N. tabacum | Cd, As | NRAMP1b metal transporter protein | Metal uptake and translocation | [36][46][47] |
| miR168 | O. sativa, | Cd | LOX1 | Promote JA synthesis | [48] |
| miR169 | M. truncatula, O. satva, N. tabaccum, B. juncea | Cr, Cd, Al, As | CCAAT-binding (TF) | ? | [35][38][44][49] |
| miR171 | M. truncatula, B. Juncea, O. sativa, N. tabacum |
Hg, As, Cr, Cd | SCL (TF) | Shoot branching, signaling pathway |
[2][35][36][50] |
| miR192 | O. sativa | Cd | ABC gene | Seed germination of rice under Cd stress | [51] |
| miR268 | O. sativa | Cd | NRAMP3 | Inhibited rice seedling growth under Cd-stress treatment | [52] |
| miR319 | M. truncatula, B. juncea, M. truncatula | Hg, As, Al | TCP (TF), cyclin | Leaf morphogenesis, cell differentiation, embryonic development, cell division |
[47][50][53] |
| miR390 | O. sativa, M. truncatula, Glycine max L. | As, Al | tasi-RNA | Plant development | [40][54][55] |
| miR393 | A. thaliana, O. sativa, M. truncatula, Saccharum spp, H. vulgare | Cd, Hg, Al | TIR1/AFBs (F -box auxin receptors) and bHLH (TF) | Regulate auxin signaling | [41][50][56][57][58] |
| miR395 | A. thaliana, B. napus, Brassica parachinensis L. | Cd, As, Al | SLIM1, SULTR2;1, APS | Sulfur assimilation, response to cadmium ion, sulfate transport |
[43][59][60][61][62] |
| miR396 | O. sativa, N. tabacum, M. sinensis, M. truncatula, G. max | As, Cr, Al | GRF (TF) | Translation, leaf development | [36][37][40][43][55] |
| miR397 | A. thaliana, O. sativa, Populus trichocarpa T. | Cu, As, Cr | Laccase | Regulate inter-tissue lignification and secondary cell wall thickness, activities of PPO, SOD, and POD | [35][59][63][64][65][66] |
| miR398 | A. thaliana, O. sativa, P. trichocarpa | Cu, Cd | CSD1, CSD2, CCS1 and COX5b-1 | SODs relive oxidative stress | [34][65][67][68] |
| miR399 | O. sativa, N. tabacum | As | ? | ? | [36][43] |
| miR408 | A. thaliana, O. sativa, P. trichocarpa, O. sava | Cu, Cr, As | Laccase, plantacyanin transcripts, SPL7, HY5 | Regulate plastocyanin (PC) content | [63] [35][43][65][69][70] |
| miR444 | O. sativa | Cr | ? | ? | [35] |
| miR528 | O. sativa, A. thaliana, Z. mays | As, Al, Cd | MAX2 gene and l-resistant blo, LACod acid oxidase, MATE, LAC | Signal transduction, regulation of cell cycle, plant development, ascorbate metabolism, miRNA processing, control of cellular-free auxin levels | [43][54][70][71][72][73] |
| miR529 | O. sativa, M. truncatula |
As, Cd | Apetala2-like (TF), squamosa promoter binding protein-like (TF) | Signaling pathway, plant development | [43][50] |
| miR808 | O. sativa, | Al | Rockweed glycosyltransferase | ? | [74] |
| miR854 | B. juncea | As | Putative serine acetyl transferase (SAT) | Synthesis of O-acetylserine | [47] |
| miR857 | A. thaliana, | Cu | Laccase (LAC7) | ? | [33] |
| miR1318 | O. sativa | As | Calcium-binding proteins or Ca2+ ATPase | Signaling | [70] |
| miR1444 | P. trichocarpa | Cu | Cu-containing proteins, polyphenol oxidases (PPOs) | ? | [65] |
| miR1535b | G. max | Cd | Glyma07g38620.1 | Responsible for initial step of isopentenyl transferase |
[75] |
In recent years, research on miRNA and HM stress has been increasing, and, with the rapid development of advanced technologies such as next-generation sequencing (NGS), a number of known and unknown miRNAs in response to metal stress are discovered through whole genome sequencing and miRNA sequencing. However, studies on the roles of different miRNAs in HMs signal transduction in plants and their targets are still in their infancy.
2. Response of miRNAs to Cu Toxicity
Cu is a trace element essential for plant growth and development. In animals, plants, and microorganisms, it is usually found in the form of Cu ions or cuprein. As an important cofactor of protein, Cu is a component of polyphenol oxidase, superoxide dismutase, laccase, cytochrome oxidase, and other enzymes, and is involved in important physiological processes such as photosynthesis, respiratory metabolism, and oxidative stress [76][77]. Cu is also a component of plastocyanin and participates in the electron transfer process of photosynthesis. However, the deficiency or excessive accumulation of Cu can cause damage to plant growth. Cu deficiency can lead to blue-green, wrinkled, distorted, or necrotic leaves, dwarf plants, slow growth, and reduced yields [78]. In contrast, excess Cu induces rapid synthesis of oxidation anions (O2−), hydroxyl radicals (OH), hydrogen peroxide (H2O2), singlet oxygen (1O2), and ROS in plant cells [79]. This reduces the strength of the cell membrane, leading to the toxic effects of Cu2+ infiltration into the cells. Excess Cu also inactivates chloroplast enzyme activity, accelerates chloroplast decomposition, inhibits chlorophyll synthesis, or rapidly compounds and destroys chlorophyll in plant cells. Additionally, Cu stress causes the electron-transport chain to be blocked, affecting plant photosynthesis [80]. Cu toxicity also affects the normal uptake of other mineral nutrients by plant roots. Cu stress disrupts the structure of protoplasts and affects their function, changing the permeability of cell membranes to increase, leading to the leakage of various ions from the membrane, and disrupting ionic equilibrium and a corresponding decrease in nutrient content [81]. To maintain the correct concentration of Cu2+ in cells, plants develop an important regulatory network in Cu uptake, distribution, and molecular responses to frequent Cu changes. The involvement of a group of miRNAs in this network appears to be particularly important, as they regulate many functionally distinct Cu proteins, including laccases, plastocyanins (PC), Cu/Zn superoxide dismutase, and polyphenol oxidase. In recent years, many studies have found that the miRNAs involved in copper stress response mainly include miR397, miR398, miR408, miR857, and miR1444, among which miR397, miR398, and miR408 are conserved in Arabidopsis and rice [32][67].
miR398 is a highly conserved miRNA in terrestrial plants, and there are three members of the miR398 family in Arabidopsis (miR398a, miR398b, miR398c) [32][67]. The sequence of miR398b and miR398c are identical, and miR398a differs from them by only one nucleotide at the 3′ end [67]. Compared with miR398b and miR398c, the promoter sequence of miR398a does not contain the GTAC sequence, which can explain the low expression of miR398a and the slow response to Cu deficiency [67]. In addition, miR398 regulates three genes through transcript cleavage and translational inhibition, including the cytosolic (CSD1) and plastidic (CSD2) genes in the Cu/Zn superoxide dismutase gene family, the copper chaperone for the superoxide dismutase (CCS1) gene [67]. The CSD1 and CSD2 genes encode closely related Cu/Zn superoxide dismutases, which detoxify superoxide free radicals. CCS1 is an intracellular Cu transporter protein with the function to transport Cu ions to the CSD through protein-to-protein interactions, which in turn activates the CSD. Under high Cu stress, the expression of miR398 suppressed, resulting in an increase in CSD1 and CSD2 expression, which relieves the threat of ROS due to the increased Cu content [67]. In the absence of Cu, the expression of miR398 would be induced, and the transcription levels of its target genes CSD1 and CSD2 would decrease, while iron superoxide dismutase (FeSOD) will be up-regulated to increase the Cu availability of other important Cu proteins (such as plastocyanin) [67]. miR398 plays a key role in the regulation of Cu homeostasis by regulating the non-essential Cu protein CSD, when Cu is lacking or excessive.
Meanwhile, miR397, miR408, and miR857 are also involved in regulating the abundance of other Cu proteins in Arabidopsis, especially the effectiveness of laccase and the secreted protein plantacyanin in response to Cu [33]. As a Cu-containing oxidase, laccase can promote the synthesis of lignin from lignin monomer in plants and promote the lignification process of plants [82]. miR397 is a key regulator of Cu homeostasis in plants such as A. thaliana, P. trichocarpa, and V. vinifera. Under low Cu stress, the expression of miR397 was up-regulated, the synthesis of laccase or plastocyanin was inhibited, and the accumulation of Cu increased, which was beneficial to maintain the homeostasis of Cu [63][64][83]. In tomato (Solanum lycopersicum M.), after overexpression of miR397a, its target gene LeLACmiR397a was down-regulated, which caused a decrease in the activities of polyphenol oxidase (PPO), superoxide dismutase (SOD), and peroxidase (POD). In addition, miR397 is also related to the yield of crops. Compared with wild-type plants, transgenic rice that overexpresses miR397 has larger seeds, increased inflorescence numbers, and increased yields [59]. Studies have shown that miR408 can respond to abiotic stress and maintain Cu homeostasis in plants [69]. A study showed that the miR408 could bind to the 5`-UTR region of LAC1, LAC12, and LAC13, and regulated the expression of these genes to maintain Cu homeostasis in plants [63][69]. It was demonstrated that transgenic strains with simultaneous inhibition of the functions of three conserved Cu-miRNAs (miR397, miR398, and miR408) showed the reduced accumulation of Cu-miRNAs, the increased accumulation of transcripts encoding Cu proteins, and that photosynthesis and growth of transgenic plants were affected under low Cu conditions, which may be associated with defective accumulation of chloroplast plastocyanin [84]. Interestingly, miR1444 is currently only found in P. trichocarpa and appears to be a Populus-specific miRNA. It has also been shown to regulate a group of Cu-containing proteins: polyphenol oxidases (PPOs) [65].
The study found that the promoter regions of these several miRNAs all have Cu-response elements (CuRE) as the role sites of Cu regulation, which is composed of repeated GTAC sequences and is the only Cu-responsive element identified to date [85]. SQUAMOSA promoter-binding protein-like-7 (SPL7) is a key transcription factor that is sensitive to low Cu and plays a transcriptional regulatory role by binding to GTAC sequences [68]. In Cu deficiency, SPL7 can activate the transcription of miR397, miR398, miR408, and miR857 genes, which inhibit the expression of copper-containing protein genes such as laccase and Cu-Zn superoxide dismutase, thereby regulating Cu in the case of Cu-deficiency allocation [68]. When Cu accumulates, SPL7 becomes inactive and the expression of miRNAs in plants are inhibited, which increases the transcription level of target genes and improves the availability of Cu proteins, thus alleviating the oxidative damage caused by copper accumulation (Figure 2).
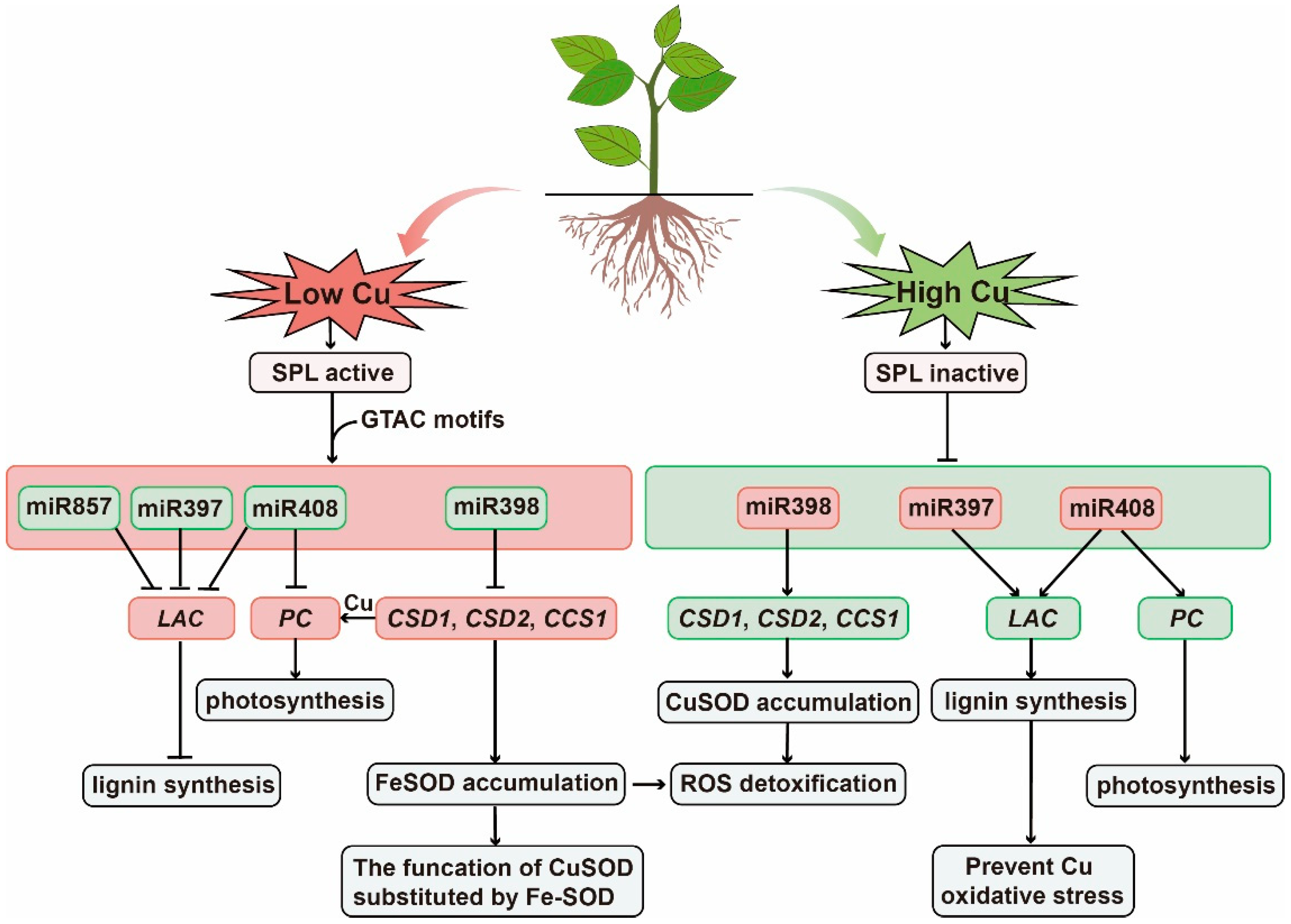
Figure 2. Schematic representation of the mode of action of miRNAs in response to Cu stress. Green boxes indicate relevant miRNAs and pathways involved in low Cu stress. SPL7 transcription factors are active and regulate miR397, miR398, miR408, and miR857, which regulate genes encoding Cu-containing proteins, including Cu/Zn superoxide dismutase (CSD), CCS1, laccase (LAC), and phycocyanin (PLC), thereby conserving Cu as essential Cu protein (such as plastocyanin). Red boxes indicate that under high Cu stress, SPL7 is inactivated, miR398 is down-regulated, and the expression of target genes CSD1, CSD2, and CCS1 is up-regulated, mitigating the threat of ROS from increased Cu content and increased Cu protein accumulation.
With the development of transcriptome and small RNA sequencing technology, a large number of studies will utilize this method to identify miRNAs in plants. In 2015, under the control and Cu stress conditions, the conservative and non-conservative miRNA and other short RNA were identified in Paeonia ostia T. [86]. A total of 102 known plant miRNAs were identified and combined with transcriptome sequencing data, while 34 new potential miRNAs were identified under the same conditions. It was also found that 12 conservative miRNA and 18 new miRNAs changed significantly under Cu stress. Jiu et al. (2019) used high-throughput sequencing to determine the miRNAs and target genes of grapevine (V. vinifera) plants responsive to Cu stress [87]. Among them, 100 known and 47 newly discovered miRNAs were differentially expressed under Cu stress. The target prediction of miRNA shows that miRNA may regulate transcription factors such as AP2, SBP, NAC, MYB, and ARF under Cu stress. They also found that miR156 targets SPL7 and may be involved in the regulation of Cu homeostasis in grapes (V. vinifera). In addition, a total of 65 known miRNAs and 78 new miRNAs predicted to mature in mulberry were identified [30]. In total, 40 miRNAs were differentially expressed under Cu stress, of which 27 miRNAs up-regulated genes and 13 miRNAs down-regulated genes. Utilizing high-throughput sequencing technology, a large number of unknown miRNAs have been found to be involved in the Cu-stress-response process. How these miRNAs regulate the distribution of Cu in plants, enabling plants to coordinate the expression and development of Cu proteins, requires further studies to explore.
References
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging Roles of MicroRNAs in Plant Heavy Metal Tolerance and Homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965.
- Gupta, O.P.; Sharma, P.; Gupta, R.K.; Sharma, I. MicroRNA Mediated Regulation of Metal Toxicity in Plants: Present Status and Future Perspectives. Plant Mol. Biol. 2014, 84, 1–18.
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front. Plant Sci. 2016, 6, 1–36.
- Cobbett, C.; Goldsbrough, P. Phytochelatins and Metallothioneins: Roles in Heavy Metal Detoxification and Homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182.
- Song, W.Y.; Park, J.; Mendoza-Cózatl, D.G.; Suter-Grotemeyer, M.; Shima, D.; Hörtensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic Tolerance in Arabidopsis Is Mediated by Two ABCC-Type Phytochelatin Transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192.
- Thomine, S.; Wang, R.; Ward, J.M.; Crawford, N.M.; Schroeder, J.I. Cadmium and Iron Transport by Members of a Plant Metal Transporter Family in Arabidopsis with Homology to Nramp Genes. Proc. Natl. Acad. Sci. USA 2000, 97, 4991–4996.
- Jaskulak, M.; Rorat, A.; Grobelak, A.; Kacprzak, M. Antioxidative Enzymes and Expression of RbcL Gene as Tools to Monitor Heavy Metal-Related Stress in Plants. J. Environ. Manage. 2018, 218, 71–78.
- Dalcorso, G.; Manara, A.; Furini, A. An Overview of Heavy Metal Challenge in Plants: From Roots to Shoots. Metallomics 2013, 5, 1117–1132.
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Biosens. Bioelectron. 2004, 116, 281–297.
- Sanz-Carbonell, A.; Marques, M.C.; Bustamante, A.; Fares, M.A.; Rodrigo, G.; Gomez, G. Inferring the Regulatory Network of the MiRNA-Mediated Response to Biotic and Abiotic Stress in Melon. BMC Plant Biol. 2019, 19, 1–17.
- Min Yang, Z.; Chen, J. A Potential Role of MicroRNAs in Plant Response to Metal Toxicity. Metallomics. 2013, 5, 1184–1190.
- Song, X.; Li, Y.; Cao, X.; Qi, Y. Annual Review of Plant Biology MicroRNAs and Their Regulatory Roles in Plant-Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 1–37.
- Jones-rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and Their Regulatory Roles in Plants. Annu. Rev. Plant Biol. 2006, 57, 19–53.
- Khraiwesh, B.; Zhu, J.; Zhu, J. Biochimica et Biophysica Acta Role of MiRNAs and SiRNAs in Biotic and Abiotic Stress Responses of Plant. Biochim. Biophys. Acta 2012, 1819, 137–148.
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S. MicroRNAs as Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 1–18.
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of MiRNA-Mediated Gene Regulation from Common Downregulation to MRNA-Specific Upregulation. Int. J. Genom. 2014, 970607.
- Gismondi, A.; Di Marco, G.; Camoni, L.; Montesano, C.; Braglia, R.; Marra, M.; Canini, A. MicroRNA Expression Profiles in Moringa Oleifera Lam. Seedlings at Different Growth Conditions. J. Plant Growth Regul. 2022, 1–9.
- Zhou, Y.; Honda, M.; Zhu, H.; Zhang, Z.; Guo, X.; Li, T.; Li, Z.; Peng, X.; Nakajima, K.; Duan, L.; et al. Spatiotemporal Sequestration of MiR165/166 by Arabidopsis Argonaute10 Promotes Shoot Apical Meristem Maintenance. Cell Rep. 2015, 10, 1819–1827.
- Knauer, S.; Holt, A.L.; Rubio-Somoza, I.; Tucker, E.J.; Hinze, A.; Pisch, M.; Javelle, M.; Timmermans, M.C.; Tucker, M.R.; Laux, T. A Protodermal MiR394 Signal Defines a Region of Stem Cell Competence in the Arabidopsis Shoot Meristem. Dev. Cell. 2013, 24, 125–132.
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by MiR319 Is Required for Compound-Leaf Development in Tomato. Nat. Genet. 2007, 39, 787–791.
- Hibara, K.I.; Karim, M.R.; Takada, S.; Taoka, K.I.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 Regulates Postembryonic Shoot Meristem and Organ Boundary Formation. Plant Cell 2006, 18, 2946–2957.
- Gutierrez, L.; Bussell, J.D.; Pǎcurar, D.I.; Schwambach, J.; Pǎcurar, M.; Bellini, C. Phenotypic Plasticity of Adventitious Rooting in Arabidopsis Is Controlled by Complex Regulation of AUXIN RESPONSE FACTOR Transcripts and MicroRNA Abundance. Plant Cell 2009, 21, 3119–3132.
- Yu, S.; Galvão, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.W. Gibberellin Regulates the Arabidopsis Floral Transition through MiR156-Targeted SQUAMOSA PROMOTER BINDING-LIKE Transcription Factors. Plant Cell 2012, 24, 3320–3332.
- Zhu, Q.H.; Helliwell, C.A. Regulation of Flowering Time and Floral Patterning by MiR172. J. Exp. Bot. 2011, 62, 487–495.
- Manavella, P.A.; Yang, S.W.; Palatnik, J. Keep Calm and Carry on: MiRNA Biogenesis under Stress. Plant J. 2019, 99, 832–843.
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat Stress Induction of MiR398 Triggers a Regulatory Loop That Is Critical for Thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851.
- Song, J.B.; Gao, S.; Wang, Y.; Li, B.W.; Zhang, Y.L.; Yang, Z.M. MiR394 and Its Target Gene LCR Are Involved in Cold Stress Response in Arabidopsis. Plant Gene. 2016, 5, 56–64.
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive Expression of a MiR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 2013, 161, 1375–1391.
- Liu, Q.; Hu, H.; Zhu, L.; Li, R.; Feng, Y. Involvement of MiR528 in the Regulation of Arsenite Tolerance in Rice (Oryza Sativa L.). Agric. Food Chem. 2015, 63, 8849–8861.
- Du, Q.; Guo, P.; Shi, Y.; Zhang, J.; Zheng, D.; Li, Y.; Acheampong, A.; Wu, P.; Lin, Q.; Zhao, W. Genome-Wide Identification of Copper Stress-Regulated and Novel MicroRNAs in Mulberry Leaf. Biochem. Genet. 2021, 59, 589–603.
- Van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538.
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced MiRNA. Mol. Cell 2004, 14, 787–799.
- Fu, Y.; Mason, A.S.; Zhang, Y.; Lin, B.; Xiao, M.; Fu, D.; Yu, H. MicroRNA-MRNA Expression Profiles and Their Potential Role in Cadmium Stress Response in Brassica Napus. BMC Plant Biol. 2019, 19, 1–20.
- Liu, Q.; Zhang, H. Molecular Identification and Analysis of Arsenite Stress-Responsive. Agric. Food Chem. 2012, 60, 6524–6536.
- Sharma, D.; Tiwari, M.; Lakhwani, D.; Tripathi, R.D.; Trivedi, P.K. Differential Expression of MicroRNAs by Arsenate and Arsenite Stress in Natural Accessions of Rice. Metallomics 2015, 7, 174–187.
- Yu, L.J.; Luo, Y.F.; Liao, B.; Xie, L.J.; Chen, L.; Xiao, S.; Li, J.T.; Hu, S.N.; Shu, W.S. Comparative Transcriptome Analysis of Transporters, Phytohormone and Lipid Metabolism Pathways in Response to Arsenic Stress in Rice (Oryza Sativa). New Phytol. 2012, 195, 97–112.
- Pandey, A.K.; Gedda, M.R.; Verma, A.K. Effect of Arsenic Stress on Expression Pattern of a Rice Specific MiR156j at Various Developmental Stages and Their Allied Co-Expression Target Networks. Front. Plant Sci. 2020, 11, 1–11.
- Shahbaz, M.; Pilon, M. Conserved Cu-MicroRNAs in Arabidopsis Thaliana Function in Copper Economy under Deficiency. Plants 2019, 8, 141.
- Xia, K.; Zeng, X.; Jiao, Z.; Li, M.; Xu, W.; Nong, Q.; Mo, H.; Cheng, T.; Zhang, M. Formation of Protein Disulfide Bonds Catalyzed by OsPDIL1;1 Is Mediated by MicroRNA5144-3p in Rice. Plant Cell Physiol. 2018, 59, 331–342.
- Cervantes, C.; Campos-García, J.; Devars, S.; Gutiérrez-Corona, F.; Loza-Tavera, H.; Torres-Guzmán, J.C.; Moreno-Sánchez, R. Interactions of Chromium with Microorganisms and Plants. FEMS Microbiol. Rev. 2001, 25, 335–347.
- Shahid, M.; Shamshad, S.; Rafiq, M.; Khalid, S.; Bibi, I.; Niazi, N.K.; Dumat, C.; Rashid, M.I. Chromium Speciation, Bioavailability, Uptake, Toxicity and Detoxification in Soil-Plant System: A Review. Chemosphere 2017, 178, 513–533.
- Conceicao Gomes, M.A.; Hauser-Davis, R.A.; Suzuki, M.S.; Vitória, A.P. Plant Chromium Uptake and Transport, Physiological Effects and Recent Advances in Molecular Investigations. Ecotoxicol. Environ. Saf. 2017, 140, 55–64.
- Azevedo, R.; Rodriguez, E. Phytotoxicity of Mercury in Plants: A Review. J. Bot. 2012, 1–6.
- Qiao, K.; Tian, Y.; Hu, Z.; Chai, T. Wheat Cell Number Regulator CNR10 Enhances the Tolerance, Translocation, and Accumulation of Heavy Metals in Plants. Environ. Sci. Technol. 2019, 53, 860–867.
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361.
- Zhou, Z.S.; Huang, S.Q.; Yang, Z.M. Bioinformatic Identification and Expression Analysis of New MicroRNAs from Medicago Truncatula. Biochem. Biophys. Res. Commun. 2008, 374, 538–542.
- Zhou, M.; Ghnaya, T.; Dailly, H.; Cui, G.; Vanpee, B.; Han, R.; Lutts, S. The Cytokinin Trans-Zeatine Riboside Increased Resistance to Heavy Metals in the Halophyte Plant Species Kosteletzkya Pentacarpos in the Absence but Not in the Presence of NaCl. Chemosphere 2019, 233, 954–965.
- Lu, S.; Yang, C.; Chiang, V.L. Conservation and Diversity of MicroRNA-Associated Copper-Regulatory Networks in Populus Trichocarpa. J. Integr. Plant Biol. 2011, 53, 879–891.
- Zhou, Z.S.; Zeng, H.Q.; Liu, Z.P.; Yang, Z.M. Genome-Wide Identification of Medicago Truncatula MicroRNAs and Their Targets Reveals Their Differential Regulation by Heavy Metal. Plant Cell Environ. 2012, 35, 86–99.
- Piotrowska-Niczyporuk, A.; Bajguz, A.; Zambrzycka, E.; Godlewska-Żyłkiewicz, B. Phytohormones as Regulators of Heavy Metal Biosorption and Toxicity in Green Alga Chlorella Vulgaris (Chlorophyceae). Plant Physiol. Biochem. 2012, 52, 52–65.
- Jin, Q.; Xue, Z.; Dong, C.; Wang, Y.; Chu, L.; Xu, Y. Identification and Characterization of MicroRNAs from Tree Peony (Paeonia Ostii) and Their Response to Copper Stress. PLoS ONE 2015, 10, 1–15.
- Gao, J.; Luo, M.; Peng, H.; Chen, F.; Li, W. Characterization of Cadmium-Responsive MicroRNAs and Their Target Genes in Maize (Zea Mays) Roots. BMC Mol. Biol. 2019, 20, 1–9.
- Tripathi, R.D.; Srivastava, S.; Mishra, S.; Singh, N.; Tuli, R.; Gupta, D.K.; Maathuis, F.J.M. Arsenic Hazards: Strategies for Tolerance and Remediation by Plants. Trends Biotechnol. 2007, 25, 158–165.
- Lièvremont, D.; Bertin, P.N.; Lett, M.C. Arsenic Behaviour in Soil-Plant System: Biogeochemical Reactions and Chemical. Speciat. Influ. 2017, 2, 97–139.
- Liu, W.; Xu, L.; Wang, Y.; Shen, H.; Zhu, X.; Zhang, K.; Chen, Y.; Yu, R.; Limera, C.; Liu, L. Transcriptome-Wide Analysis of Chromium-Stress Responsive MicroRNAs to Explore MiRNA-Mediated Regulatory Networks in Radish (Raphanus Sativus L.). Sci. Rep. 2015, 5, 1–17.
- Ding, Y.; Wang, Y.; Jiang, Z.; Wang, F.; Jiang, Q.; Sun, J.; Chen, Z.; Zhu, C. MicroRNA268 Overexpression Affects Rice Seedling Growth under Cadmium Stress. J. Agric. Food Chem. 2017, 65, 5860–5867.
- Gill, R.A.; Zang, L.; Ali, B.; Farooq, M.A.; Cui, P.; Yang, S.; Ali, S.; Zhou, W. Chromium-Induced Physio-Chemical and Ultrastructural Changes in Four Cultivars of Brassica Napus L. Chemosphere 2015, 120, 154–164.
- Bukhari, S.A.H.; Shang, S.; Zhang, M.; Zheng, W.; Zhang, G.; Wang, T.Z.; Shamsi, I.H.; Wu, F. Genome-Wide Identification of Chromium Stress-Responsive Micro RNAs and Their Target Genes in Tobacco (Nicotiana Tabacum) Roots. Environ. Toxicol. Chem. 2015, 34, 2573–2582.
- Min, H.L.; Cai, S.J.; Rui, Z.; Sha, S.; Xie, K.B.; Xu, Q.S. Calcium-Mediated Enhancement of Copper Tolerance in Elodea Canadensis. Biol. Plant 2013, 57, 365–369.
- Ding, Y.; Qu, A.; Gong, S.; Huang, S.; Lv, B.; Zhu, C. Molecular Identification and Analysis of Cd-Responsive MicroRNAs. Agric. Food Chem. 2013, 61, 11668–11675.
- Chen, L.; Wang, T.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of Aluminum-Responsive MicroRNAs in Medicago Truncatula by Genome-Wide High-Throughput Sequencing. Planta 2012, 235, 375–386.
- Iwase, J.; Furukawa, H.; Hiramatsu, T.; Bouteau, F.; Mancuso, S.; Tanaka, K.; Okazaki, T.; Kawano, T. Protection of Tobacco Cells from Oxidative Copper Toxicity by Catalytically Active Metal-Binding DNA Oligomers. J. Exp. Bot. 2014, 65, 1391–1402.
- Obrist, D.; Kirk, J.L.; Zhang, L.; Sunderland, E.M.; Jiskra, M.; Selin, N.E. A Review of Global Environmental Mercury Processes in Response to Human and Natural Perturbations: Changes of Emissions, Climate, and Land Use. Ambio 2018, 47, 116–140.
- Demidchik, V.; Sokolik, A.; Yurin, V. The Effect of Cu2+ on Ion Transport Systems of the Plant Cell Plasmalemma. Plant Physiol. 1997, 114, 1313–1325.
- Pegler, J.L.; Oultram, J.M.J.; Nguyen, D.Q.; Grof, C.P.L.; Eamens, A.L. Microrna-Mediated Responses to Cadmium Stress in Arabidopsis Thaliana. Plants 2021, 10, 130.
- Pilon, M.; Abdel-Ghany, S.E.; Cohu, C.M.; Gogolin, K.A.; Ye, H. Copper Cofactor Delivery in Plant Cells. Curr. Opin. Plant Biol. 2006, 9, 256–263.
- Heaton, A.C.; Rugh, C.L.; Wang, N.J.; Meagher, R.B. Physiological Responses of Transgenic merA-TOBACCO (Nicotiana tabacum) to Foliar and Root Mercury Exposure. Water Air Soil Pollut. 2005, 161, 137–155.
- Nie, G.; Liao, Z.; Zhong, M.; Zhou, J.; Cai, J.; Liu, A.; Wang, X.; Zhang, X. MicroRNA-Mediated Responses to Chromium Stress Provide Insight Into Tolerance Characteristics of Miscanthus Sinensis. Front. Plant Sci. 2021, 12.
- Ye, Z.; Zeng, J.; Long, L.; Ye, L.; Zhang, G. Identification of MicroRNAs in Response to Low Potassium Stress in the Shoots of Tibetan Wild Barley and Cultivated. Curr. Plant Biol. 2021, 25, 1–14.
- Kopittke, P.M.; Moore, K.L.; Lombi, E.; Gianoncelli, A.; Ferguson, B.J.; Blamey, F.P.C.; Menzies, N.W.; Nicholson, T.M.; McKenna, B.A.; Wang, P.; et al. Identification of the Primary Lesion of Toxic Aluminum in Plant Roots. Plant Physiol. 2015, 167, 1402–1411.
- Silva, S. Aluminium Toxicity Targets in Plants. J. Bot. 2012, 2012, 1–8.
- Yuan, N.; Yuan, S.; Li, Z.; Li, D.; Hu, Q.; Luo, H. Heterologous Expression of a Rice MiR395 Gene in Nicotiana Tabacum Impairs Sulfate Homeostasis. Sci. Rep. 2016, 6, 1–14.
- Vinit-Dunand, F.; Epron, D.; Alaoui-Sossé, B.; Badot, P.M. Effects of Copper on Growth and on Photosynthesis of Mature and Expanding Leaves in Cucumber Plants. Plant Sci. 2002, 163, 53–58.
- Pätsikkä, E.; Kairavuo, M.; Šeršen, F.; Aro, E.M.; Tyystjärvi, E. Excess Copper Predisposes Photosystem II to Photoinhibition in Vivo by Outcompeting Iron and Causing Decrease in Leaf Chlorophyll. Plant Physiol. 2002, 129, 1359–1367.
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of MiR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065.
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-Throughput Sequencing of Arabidopsis MicroRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS ONE 2007, 2, e219.
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. LACCASE Is Necessary and Nonredundant with PEROXIDASE for Lignin Polymerization during Vascular Development in Arabidopsis. Plant Cell 2013, 25, 3976–3987.
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-Mediated Systemic down-Regulation of Copper Protein Expression in Response to Low Copper Availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945.
- Wang, C.Y.; Zhang, S.; Yu, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X.; et al. MiR397b Regulates Both Lignin Content and Seed Number in Arabidopsis via Modulating a Laccase Involved in Lignin Biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142.
- Leng, X.; Wang, P.; Zhao, P.; Wang, M.; Cui, L.; Shangguan, L.; Wang, C.; Wang, C. Conservation of MicroRNA-Mediated Regulatory Networks in Response to Copper Stress in Grapevine. Plant Growth Regul. 2017, 82, 293–304.
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.J.; Qu, L.H.; Chen, F.; et al. Overexpression of MicroRNA OsmiR397 Improves Rice Yield by Increasing Grain Size and Promoting Panicle Branching. Nat. Biotechnol. 2013, 31, 848–852.
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. Microrna408 Is Critical for the HY5-SPl7 Gene Network That Mediates the Coordinated Response to Light and Copper. Plant Cell 2014, 26, 4933–4953.
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. Toxicity Responses of Cu and Cd: The Involvement of MiRNAs and the Transcription Factor SPL7. BMC Plant Biol. 2016, 16, 1–16.
- Jiu, S.; Leng, X.; Haider, M.S.; Dong, T.; Guan, L.; Xie, Z.; Li, X.; Shangguan, L.; Fang, J. Identification of Copper (Cu) Stress-Responsive Grapevine MicroRNAs and Their Target Genes by High-Throughput Sequencing. R. Soc. Open Sci. 2019, 6, 180735.
- Xu, L.; Wang, Y.; Liu, W.; Wang, J.; Zhu, X.; Zhang, K.; Yu, R.; Wang, R.; Xie, Y.; Zhang, W.; et al. De Novo Sequencing of Root Transcriptome Reveals Complex Cadmium-Responsive Regulatory Networks in Radish (Raphanus Sativus L.). Plant Sci. 2015, 236, 313–323.
- Gu, Q.; Chen, Z.; Cui, W.; Zhang, Y.; Hu, H.; Yu, X.; Wang, Q.; Shen, W. Methane Alleviates Alfalfa Cadmium Toxicity via Decreasing Cadmium Accumulation and Reestablishing Glutathione Homeostasis. Ecotoxicol. Environ. Saf. 2018, 147, 861–871.
- Ding, Y.F.; Zhu, C. The Role of MicroRNAs in Copper and Cadmium Homeostasis. Biochem. Biophys. Res. Commun. 2009, 386, 6–10.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
838
Revisions:
2 times
(View History)
Update Date:
20 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




