Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chen, H.; Zhou, X.; Wang, M.; Ren, L. CRISPR/Cas-Based Detection. Encyclopedia. Available online: https://encyclopedia.pub/entry/41236 (accessed on 03 March 2026).
Chen H, Zhou X, Wang M, Ren L. CRISPR/Cas-Based Detection. Encyclopedia. Available at: https://encyclopedia.pub/entry/41236. Accessed March 03, 2026.
Chen, Haoxiang, Xi Zhou, Miao Wang, Lei Ren. "CRISPR/Cas-Based Detection" Encyclopedia, https://encyclopedia.pub/entry/41236 (accessed March 03, 2026).
Chen, H., Zhou, X., Wang, M., & Ren, L. (2023, February 15). CRISPR/Cas-Based Detection. In Encyclopedia. https://encyclopedia.pub/entry/41236
Chen, Haoxiang, et al. "CRISPR/Cas-Based Detection." Encyclopedia. Web. 15 February, 2023.
Copy Citation
Rapid, accurate, and portable on-site detection is critical in the face of public health emergencies. Infectious disease control and public health emergency policymaking can both be aided by effective and trustworthy point of care tests (POCT). A very promising POCT method appears to be the clustered regularly interspaced short palindromic repeats and associated protein (CRISPR/Cas)-based molecular diagnosis. For on-site detection, CRISPR/Cas-based detection can be combined with multiple signal sensing methods and integrated into smart devices.
CRISPR/Cas
molecular diagnostic
DNA detection
point of care test
1. Introduction
Nucleic acid-based tests have emerged as the gold standard for many diseases, including pathogenic infections, despite the fact that there are various forms of clinical diagnosis. Quantitative polymerase chain reaction (qPCR)-based nucleic acid testing is also widely employed in many fields including public health, food safety, environmental monitoring, etc., [1][2]. Because this method relies on specialist instruments and operators, point of care testing (POCT), which is instrument-independent and user-friendly, has received a lot of attention, especially during the global SARS-CoV-2 pandemic. In recent years, numerous technologies have been developed and applied to molecular diagnostics, such as nanotechnology, nucleic acid amplification, interface modification and sensing, and molecular assembly techniques [3][4][5]. In particular, CRISPR-based diagnostics have been hailed as a promising candidate for next-generation diagnostics because of their capacity to detect nucleic acids rapidly and accurately, without relying on technical expertise and assistive equipment [6].
CRISPR/Cas systems are the adaptive immune system of bacteria, which can resist the invasion of exogenous genes [7][8]. CRISPR/Cas systems consist of the CRISPR array and CRISPR-associated protein (Cas protein). The CRISPR array contains repeats and spacer sequences to generate CRISPR RNA (crRNA), which combines with the corresponding Cas protein to form a ribonucleoprotein (RNP) complex that functions as a genomic DNA editor [9]. CRISPR/Cas systems have been divided into two classes, and class 2 systems are widely used because they require only one Cas protein (e.g., Cas9, Cas12a, Cas13a) to fulfill their corresponding functions [10][11]. Since the team led by Emmanuelle Charpentier and Jennifer A. Doudna clarified the DNA-editing mechanism of the class 2 CRISPR system, genome engineering has gained tremendous momentum [8][11]. With the comprehensive understanding of CRISPR/Cas biology, the indiscriminate trans-cleavage activity of Cas12a and Cas13a has been reported and can be used in combination with single-strand DNA (ssDNA) or single-strand RNA (ssRNA) probes for the development of molecular diagnostic methods [12][13][14]. After target cleavage, the Cas9 protein goes into an inactive state, but when the Cas12a and Cas13a proteins are activated, indiscriminate trans-cleavage activity is maintained, in stark contrast to Cas9. Due to this unique property, both the Cas12a and Cas13a proteins can be triggered by a single target sequence to acquire multiple nuclease activities. These activities significantly increase sensitivity by acting as superior signal amplifiers for nucleic acid detection.
2. Sensing Methods for CRISPR/Cas-Based Detection
2.1. Fluorescence Signal Sensing
After activation, both Cas12a and Cas13a proteins show a trans-cleavage activity and can cleave single-stranded DNA (ssDNA) and single-stranded RNA (ssRNA) nonspecifically, respectively [12][13]. Upon the recognition of the target sequence by Cas12a or Cas13a, the Cas effector proteins can indiscriminately cleave short ssDNA or ssRNA bearing both a fluorophore and a quencher, reporting a fluorescent signal [15][16]. CRISPR-based nucleic acid diagnosis is usually divided into four steps. Firstly, nucleic acid extraction. Commercial kits can be used to swiftly extract the target nucleic acid in clinical samples such as blood, urine, and nose/throat swabs, etc. Secondly, nucleic acid amplification. Isothermal amplification is frequently used for the second step. Thirdly, CRISPR-based detection, with CRISPR system selection according to different nucleic acid types. Finally, signal output. Fluorescence signals are common CRISPR sensing signals that facilitate analysis. The viral genomic nucleic acid detection limits of the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) and the DNA endonuclease-targeted CRISPR trans reporter (DETECTR) developed following this principle, are at the aM level [12][17]. CRISPR diagnosis based on fluorescence signal has great advantages in responding to public health emergencies (such as SARS-CoV-2), especially when combined with isothermal amplification technology, which can rapidly detect infectious disease pathogen nucleic acids under isothermal conditions, and only requires a portable light-emitting diode (LED) device to visualize detection results [18]. Meanwhile, isothermal amplification and CRISPR-based diagnostics can be integrated into one pot, avoiding the aerogel contamination of pathogenic nucleic acids, while also reducing detection costs (Figure 1A) [19][20]. The CRISPR detection outcomes based on fluorescence signals are generally simple to see with the naked eye, requiring no specialized analytical tools and significantly lowering the cost of CRISPR-based POCT.
To employ CRISPR/Cas-based diagnostics in the field, a lateral flow assay was combined with the CRISPR/Cas system to create a convenient detection platform [15][21]. Typically, carboxyfluorescein (FAM)-biotin reporters are used to carrying out lateral flow strip-based CRISPR/Cas12a detection assays. The first detection line (control-line) captures undegraded reporters, whereas the second detection line produces a signal from indiscriminate Cas12 trans-cleavage activity (test-line) [21]. Contrarily, in a different approach, the biotinylated ssDNA reporters are degraded by Cas12a in the presence of the target, making it invisible on the test-line of a lateral flow strip since it is unable to connect to DNA probes that have already been fixed there (Figure 1B). By using a one-pot method that combines genome release, isothermal amplification, and CRISPR/Cas detection, the nucleic acid detection of pathogens can be accomplished relatively quickly. The CRISPR/Cas-cleaved fluorescent molecules can be sensitively excited by the LED, enabling visualization of the detection signal. Meanwhile, this operationally friendly detection mode has been applied in a variety of pathogen detection, such as SARS-CoV-2, Plasmodium, and African swine-fever viruses, which makes it a convenient tool for public health detection in resource-constrained areas and on-site detection [22][23][24][25].
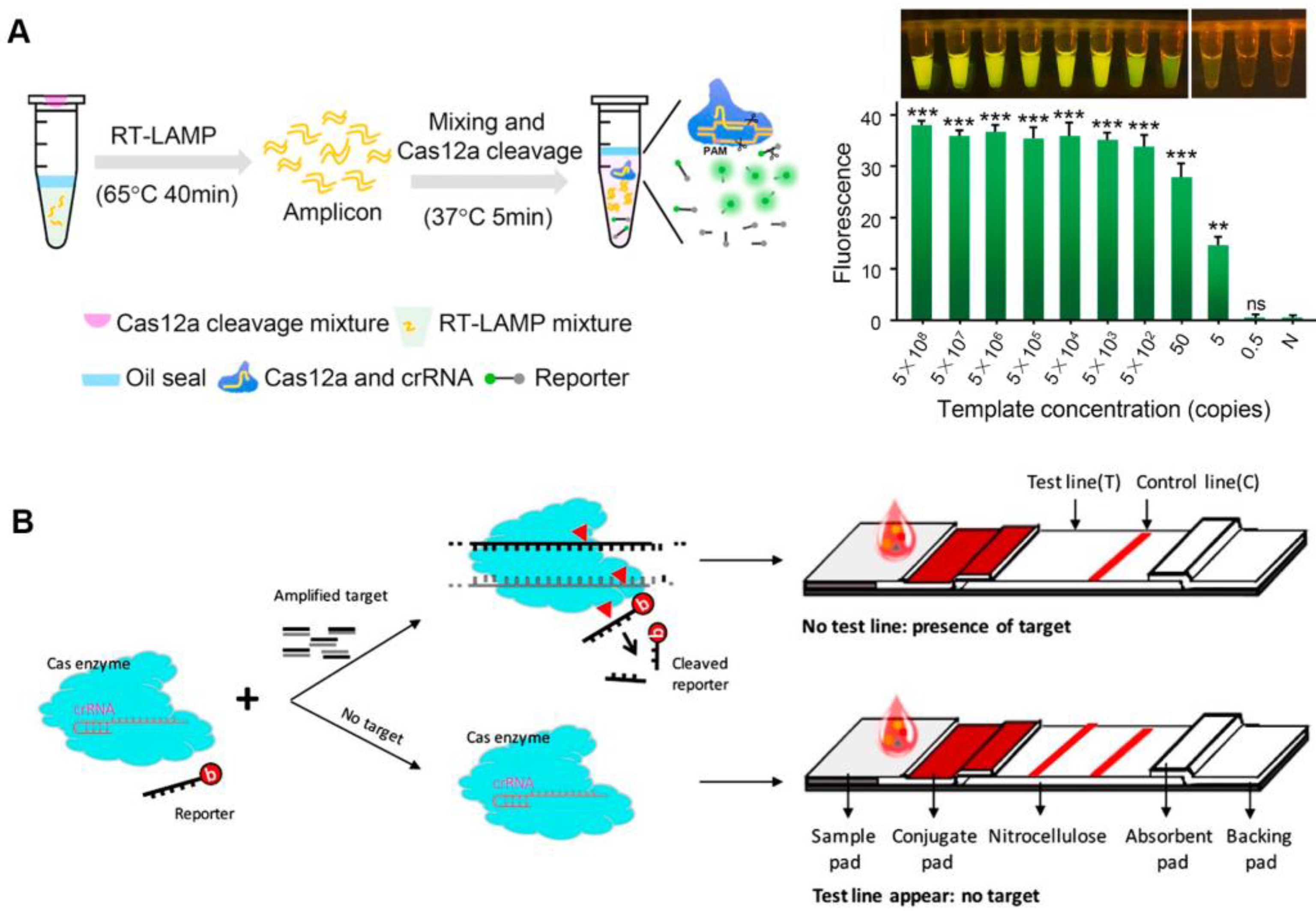

Figure 1. Fluorescence sensing strategy for CRISPR-based diagnosis. (A) The scheme of one-pot visual CRISPR-based (opvCRISPR) detection method. (ns = no significant, ** p < 0.01, *** p < 0.001) (B) Schematic diagram of CRISPR/Cas-based lateral flow assay.
2.2. Electrical Signal Sensing
The high sensitivity of CRISPR/Cas-based detection is typically based on isothermal amplification that can amplify nucleic acid molecules; however, the use of the isothermal amplification reagents raises the cost of CRISPR/Cas-based diagnosis. Because of its high sensitivity, electrical signal sensing could be a candidate for amplification-free CRISPR/Cas-based diagnosis [26][27]. Electrical signal-based CRISPR/Cas diagnosis is typically performed using a defective Cas protein such as dCas9, which can only bind and not cleave target nucleic acid molecules. Hajian et al. fixed the CRISPR/dCas9 to a field effect transistor (gFET), and when the RNP recognized and bound a target nucleic acid molecule, the gFET conductivity changed, therefore reporting the detection result [28]. Their sensor is based on the idea that charged DNA molecules can be captured by dCas9 immobilized on graphene, which lowers the system’s resistance and results in a larger current response. To prevent the nonspecific adsorption of charged molecules, the remaining graphene surface was blocked. A drop of the sample was applied to the apparatus while it was in use; it was left while it was incubated, washed, and eventually had the electrical conductivity of the platform evaluated by two platinum electrodes. The method has a detection limit of 15 fM without amplification, and the detection time is about 15 min. The nanopore sensor can also be used as a signal sensing method for CRISPR/dCas9-based diagnostic by detecting the characteristic blockade signal of dCas9-DNA as it passes through the nanopore [29]. The nanopore signals (ion current) for different dCas9-binding sites can be resolved, which could lead to a multiplexing strategy. Signal barcodes can be created to distinguish the different sequences of the DNA using different crRNAs, which can bind to different sites on the same DNA (Figure 2A) [30]. This method’s ability to discriminate between different DNA sequences in DNA mixture demonstrates its specificity and potential for multiplexing. Although dCas9 can detect and type single DNA, its application is limited by low throughput. Additionally, because there is a decreased probability of the target DNA strand passing through the nanopore in samples containing less target DNA, a longer measuring time is needed. The trans-cleavage of Cas12a may be able to provide a solution to this issue. The Cas12a trans-cleavage can degrade the DNA probes of nanopore, causing the electrical signal of the DNA probes to change significantly. The Cas12a-based nanopore sensing technology can amplify the detection signal, using its trans-cleavage activity to achieve rapid and sensitive detection of HIV-1 and SARS-CoV-2 (Figure 2B) [31][32].
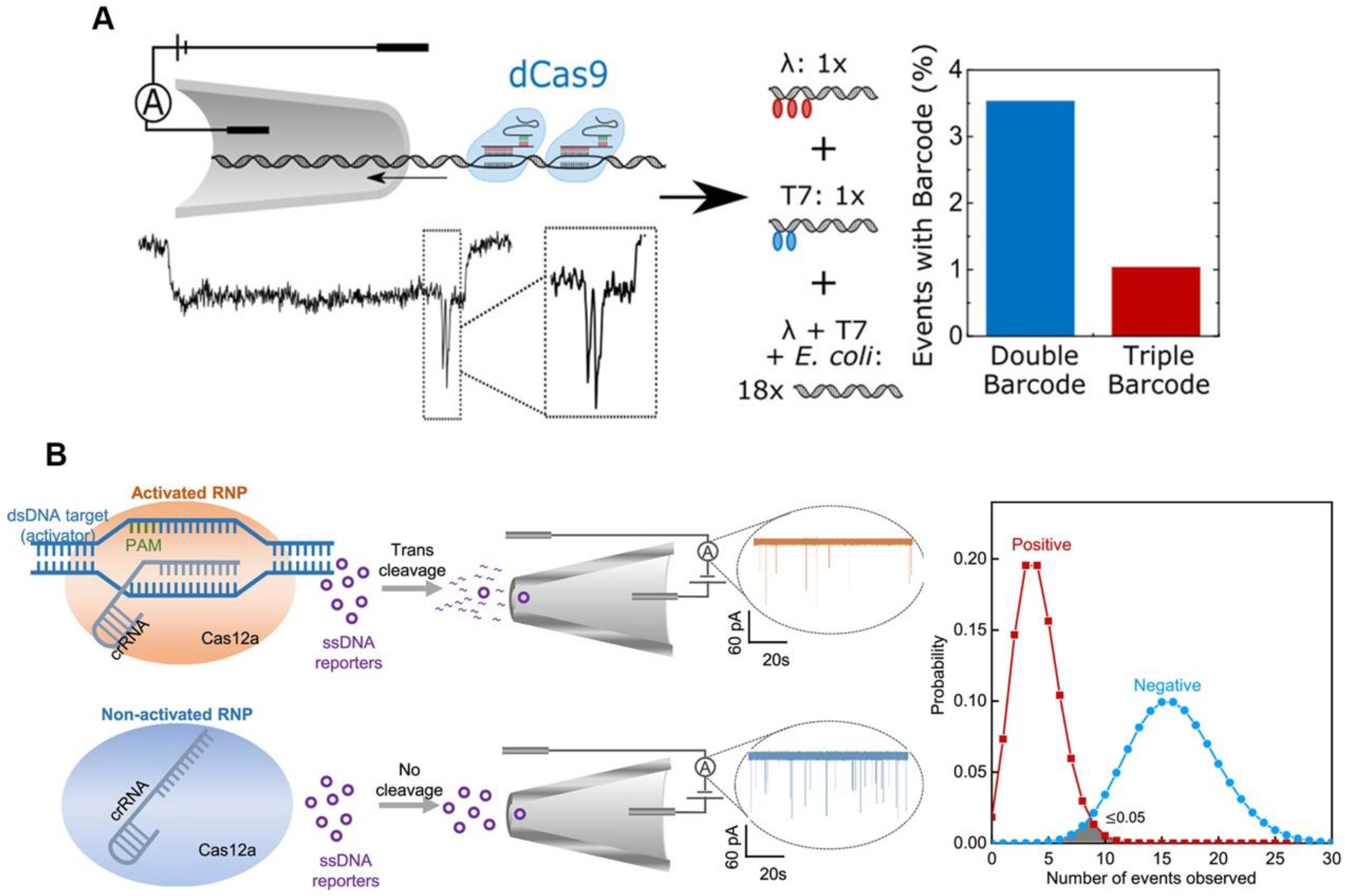
Figure 2. Electrical sensing strategy for CRISPR-based diagnosis. (A) Schematic diagram and results of nanopore-based CRISPR/dCas9 test. (B) Schematic diagram and results of nanopore-based CRISPR/Cas12a assay.
2.3. Electrochemical Signal Sensing
Methylene blue (MB) is a classic electrochemical tag, and the MB-based CRISPR diagnostic technique has high sensitivity. The ssDNA-MB was immobilized onto the gold electrode surface via sulfhydryl groups, and the MB was released when the ssDNA was trans-cleaved after the CRISPR/Cas12a was activated by the target sequences. The detection signal was reported using the current peak of the electrode measured by square wave voltammetry (SWV), which decreases with the release of MB [33]. The use of hairpin ssDNA allows MB tags to be positioned closer to the electrode surface, resulting in larger initial current peaks and more significant current peak changes after CRISPR/Cas12a-based detection (Figure 3A) [34]. The strategy of removing electrochemical tags from the electrode surface after the CRISPR/Cas system is activated leads to significant changes in the electrode’s electrochemical signal, which appears to be a promising route of CRISPR-based POCT [35].
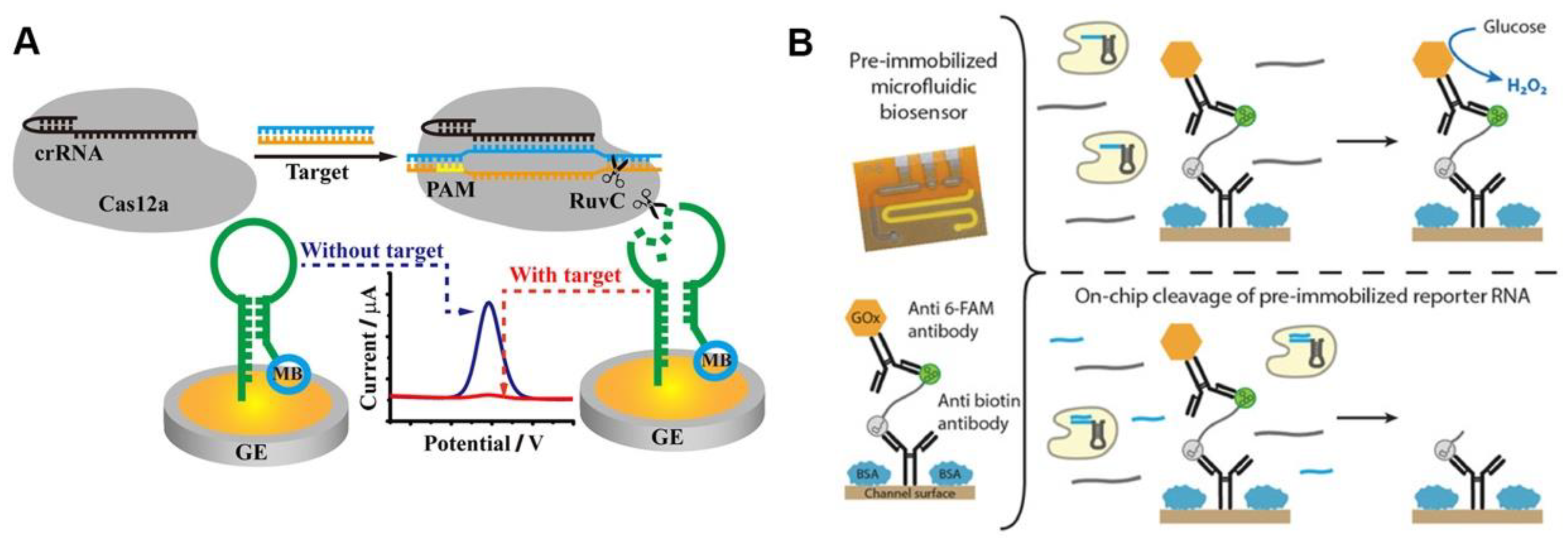
Figure 3. Electrochemical sensing for CRISPR-based diagnosis. (A) A schematic diagram of the CRISPR biosensing method based on electrochemical label. (B) A working principle of glucose oxidase-based electrochemical CRISPR assay.
The use of oxidative enzymes to amplify electrical signals appears to be a promising strategy. Bruch et al. immobilized glucose oxidase on the electrode surface using ssRNA, and the trans-cleavage activity of Cas13a cleaved ssRNA after being activated by the target sequence, releasing the glucose oxidase (Figure 3B). Glucose oxidase can oxidize the glucose in a solution to produce D-glucose-δ-lactones and H2O2, which can be measured amperometrically. The current signal will be lower than the blank if target RNA is present [36]. Additionally, independent of the electrochemical tag, the use of electrochemical impedance spectroscopy (EIS) as the detection signal may improve the electrochemical CRISPR-based diagnostic sensitivity. The electrode surface was modified with ssDNA, which was degraded after the CRISPR/Cas12a system recognized the target sequence, and the electrochemical impedance spectral transition of the electrode was reported as the detection signal [37]. In summary, the electrochemical CRISPR/Cas-based assay has achieved a highly sensitive detection of pathogens such as human papillomavirus 16 (HPV-16), human immunodeficiency virus (HIV), parvovirus B19 (PB-19), and Listeria monocytogenes, with great potential for POCT of infectious diseases [33][38][39].
The use of oxidative enzymes to amplify electrical signals appears to be a promising strategy. Bruch et al. immobilized glucose oxidase on the electrode surface using ssRNA, and the trans-cleavage activity of Cas13a cleaved ssRNA after being activated by the target sequence, releasing the glucose oxidase (Figure 3B). Glucose oxidase can oxidize the glucose in a solution to produce D-glucose-δ-lactones and H2O2, which can be measured amperometrically. The current signal will be lower than the blank if target RNA is present [36]. Additionally, independent of the electrochemical tag, the use of electrochemical impedance spectroscopy (EIS) as the detection signal may improve the electrochemical CRISPR-based diagnostic sensitivity. The electrode surface was modified with ssDNA, which was degraded after the CRISPR/Cas12a system recognized the target sequence, and the electrochemical impedance spectral transition of the electrode was reported as the detection signal [37]. In summary, the electrochemical CRISPR/Cas-based assay has achieved a highly sensitive detection of pathogens such as human papillomavirus 16 (HPV-16), human immunodeficiency virus (HIV), parvovirus B19 (PB-19), and Listeria monocytogenes, with great potential for POCT of infectious diseases [33][38][39].
2.4. Colorimetric Signal Sensing
Colorimetric sensors are popular because they can read the detection results directly with the naked eye, and colorimetric lateral flow assay (LFA) also has many applications in CRISPR/Cas-based detection (Figure 4A). The sample pad, control-line (C-line), and test-line (T-line) of commercial universal test strips were precoated with gold particle-bound anti-FAM antibodies, biotin ligands, and anti-rabbit antibodies, respectively. Biotin- and FAM-modified ssDNA served as reporters for CRISPR/Cas12a-based diagnostics, the reporters were cleaved and captured by T-line when the CRISPR/Cas system was activated, and a positive result was reported. Additionally, a negative result was reported when the reporters were captured by C-line [40]. A similar strategy is used for the on-site detection of Pseudomonas aeruginosa, Leptosphaeria maculans, and African swine fever virus, with miniaturized equipment and friendly operations flexibly applied in different fields; it is a promising POCT candidate for response to public health events [41][42][43]. Although the LFA-based CRISPR diagnosis is challenging to quantify, its benefits of rapid detection and high sensitivity have significant field application value. Furthermore, LFA sensing methods are also available for Cas9-based diagnostics. After isothermal amplification, the target sequence can be introduced to biotin by biotin-modified primers, while being bound to the avidin-modified T-line. The gold nanoparticles labeled dCas9/crRNA complexes via surface binding ssDNA, which can partially hybridize with crRNA, similar to gold nanoparticle labeled nucleic acid antibodies [44]. Another Cas9-based LFA assay also similarly requires the introduction of special chemical groups (digoxin, biotin, and fluorescein isothiocyanate isomer (FITC)) to the target sequence during isothermal processing using primers with specific chemical modifications. The Cas9nAR specifically selects isothermal amplification sites, and chemically modified primers introduce specific groups to the amplicon that can be captured by the T-line [45].
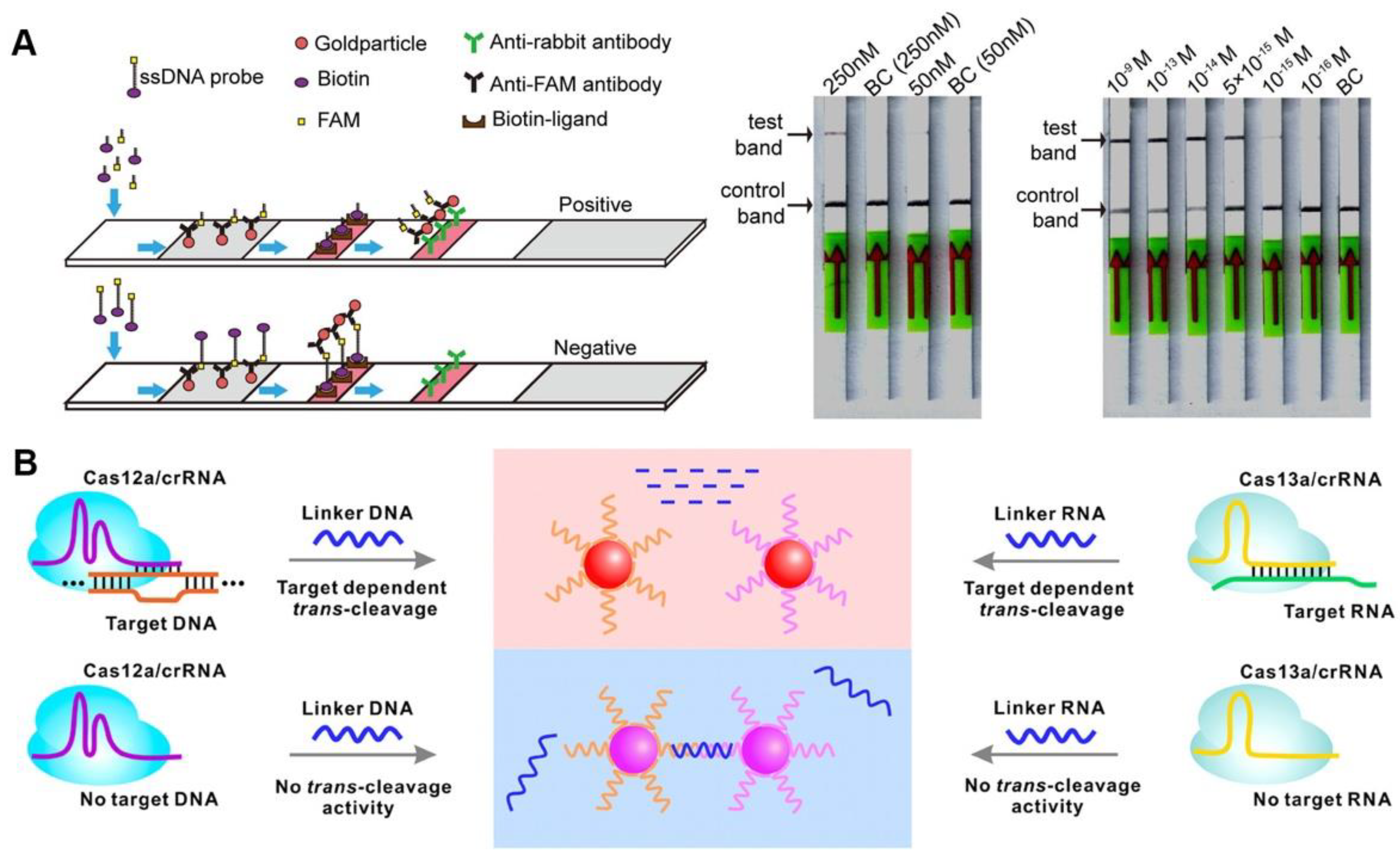
Figure 4. CRISPR-based colorimetric detection. (A) A schematic diagram and results of CORDS test. (B) A schematic illustration of gold nanoparticles-based colorimetric CRISPR assay.
References
- Dewald, F.; Suárez, I.; Johnen, R.; Grossbach, J.; Moran-Tovar, R.; Steger, G.; Joachim, A.; Rubio, G.H.; Fries, M.; Behr, F.; et al. Effective high-throughput RT-qPCR screening for SARS-CoV-2 infections in children. Nat. Commun. 2022, 13, 3640.
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108.
- Zhou, H.; Liu, J.; Xu, J.-J.; Zhang, S.; Chen, H.-Y. Advances in DNA/RNA detection using nanotechnology. Adv. Clin. Chem. 2019, 91, 31–98.
- Zhou, H.; Liu, J.; Xu, J.-J.; Zhang, S.-S.; Chen, H.-Y. Optical nano-biosensing interface via nucleic acid amplification strategy: Construction and application. Chem. Soc. Rev. 2018, 47, 1996–2019.
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545.
- Chertow, D.S. Next-generation diagnostics with CRISPR. Science 2018, 360, 381–382.
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61.
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182.
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 2014, 343, 1247997.
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c. Science 2017, 356, 438–442.
- Fonfara, I.; Richter, H.; Bratovič, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521.
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Sci. 2018, 360, 439–444.
- Cai, X.; Zhao, D.; Li, X.; Zheng, Q.; Shu, X.; Ding, S.; Zhang, D.; Yan, Y. An ultrasensitive biosensing platform for FEN1 activity detection based on target-induced primer extension to trigger the collateral cleavage of CRISPR/Cas12a. Anal. Chim. Acta 2022, 1233, 340519.
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448.
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494.
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-CoV-2 detection. Biosens. Bioelectron. 2021, 172, 112766.
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-19. Nat. Biotechnol. 2020, 38, 870–874.
- Yu, S.; Nimse, S.B.; Kim, J.; Song, K.S.; Kim, T. Development of a Lateral Flow Strip Membrane Assay for Rapid and Sensitive Detection of the SARS-CoV-19. Anal. Chem. 2020, 92, 14139–14144.
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew. Chem. Int. Ed. 2021, 60, 5307–5315.
- Lee, R.A.; De Puig, H.; Nguyen, P.Q.; Angenent-Mari, N.M.; Donghia, N.M.; McGee, J.P.; Dvorin, J.D.; Klapperich, C.M.; Pollock, N.R.; Collins, J.J. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. USA 2020, 117, 25722–25731.
- Wu, J.; Mukama, O.; Wu, W.; Li, Z.; De Dieu Habimana, J.; Zhang, Y.; Zeng, R.; Nie, C.; Zeng, L. A CRISPR/Cas12a Based Universal Lateral Flow Biosensor for the Sensitive and Specific Detection of African Swine-Fever Viruses in Whole Blood. Biosensors 2020, 10, 203.
- Swetha, P.D.P.; Sonia, J.; Sapna, K.; Prasad, K.S. Towards CRISPR powered electrochemical sensing for smart diagnostics. Curr. Opin. Electrochem. 2021, 30, 100829.
- Sun, Y.; Yang, C.; Jiang, X.; Zhang, P.; Chen, S.; Su, F.; Wang, H.; Liu, W.; He, X.; Chen, L.; et al. High-intensity vector signals for detecting SARS-CoV-2 RNA using CRISPR/Cas13a couple with stabilized graphene field-effect transistor. Biosens. Bioelectron. 2022, 222, 114979.
- Hajian, R.; Balderston, S.; Tran, T.; Deboer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437.
- Yang, W.; Restrepo-Perez, L.; Bengtson, M.; Heerema, S.J.; Birnie, A.; van der Torre, J.; Dekker, C. Detection of CRISPR-dCas9 on DNA with Solid-State Nanopores. Nano Lett. 2018, 18, 6469–6474.
- Weckman, N.E.; Ermann, N.; Gutierrez, R.; Chen, K.; Graham, J.; Tivony, R.; Heron, A.; Keyser, U.F. Multiplexed DNA Identification Using Site Specific dCas9 Barcodes and Nanopore Sensing. ACS Sens. 2019, 4, 2065–2072.
- Nouri, R.; Jiang, Y.; Lian, X.L.; Guan, W. Sequence-Specific Recognition of HIV-1 DNA with Solid-State CRISPR-Cas12a-Assisted Nanopores (SCAN). ACS Sensors 2020, 5, 1273–1280.
- Nouri, R.; Jiang, Y.; Tang, Z.; Lian, X.L.; Guan, W. Detection of SARS-CoV-2 with solid-state CRISPR-Cas12a-assisted nanopores. Nano Lett. 2021, 21, 8393–8400.
- Dai, Y.; Somoza, R.; Wang, L.; Welter, J.F.; Li, Y.; I Caplan, A.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. 2019, 58, 17399–17405.
- Zhang, D.; Yan, Y.; Que, H.; Yang, T.; Cheng, X.; Ding, S.; Zhang, X.; Cheng, W. CRISPR/Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing. ACS Sensors 2020, 5, 557–562.
- Xu, W.; Jin, T.; Dai, Y.; Liu, C.C. Surpassing the detection limit and accuracy of the electrochemical DNA sensor through the application of CRISPR Cas systems. Biosens. Bioelectron. 2020, 155, 112100.
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, e1905311.
- Bonini, A.; Poma, N.; Vivaldi, F.; Biagini, D.; Bottai, D.; Tavanti, A.; Di Francesco, F. A label-free impedance biosensing assay based on CRISPR/Cas12a collateral activity for bacterial DNA detection. J. Pharm. Biomed. Anal. 2021, 204, 114268.
- Zamani, M.; Robson, J.M.; Fan, A.; Bono, M.S.; Furst, A.L.; Klapperich, C.M. Electrochemical Strategy for Low-Cost Viral Detection. ACS Central Sci. 2021, 7, 963–972.
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Zhang, J.; Pang, R.; Xue, L.; Wang, J.; Zeng, H.; Wu, S.; et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021, 179, 113073.
- Bai, J.; Lin, H.; Li, H.; Zhou, Y.; Liu, J.; Zhong, G.; Wu, L.; Jiang, W.; Du, H.; Yang, J.; et al. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Front. Microbiol. 2019, 10, 2830.
- Mukama, O.; Wu, J.; Li, Z.; Liang, Q.; Yi, Z.; Lu, X.; Liu, Y.; Liu, Y.; Hussain, M.; Makafe, G.G.; et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020, 159, 112143.
- Lei, R.; Li, Y.; Li, L.; Wang, J.; Cui, Z.; Ju, R.; Jiang, L.; Liao, X.; Wu, P.; Wang, X. A CRISPR/Cas12a-based portable platform for rapid detection of Leptosphaeria maculans in Brassica crops. Front. Plant Sci. 2022, 13, 976510.
- Wang, X.; Ji, P.; Fan, H.; Dang, L.; Wan, W.; Liu, S.; Li, Y.; Yu, W.; Li, X.; Ma, X.; et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020, 3, 62.
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Wang, H.; Zhang, G.; Sun, J.; Zhou, X. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Lateral Flow Nucleic Acid Assay. ACS Nano 2020, 14, 2497–2508.
- Wang, L.; Shen, X.; Wang, T.; Chen, P.; Qi, N.; Yin, B.-C.; Ye, B.-C. A lateral flow strip combined with Cas9 nickase-triggered amplification reaction for dual food-borne pathogen detection. Biosens. Bioelectron. 2020, 165, 112364.
More
Information
Subjects:
Chemistry, Analytical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.5K
Revisions:
2 times
(View History)
Update Date:
15 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





