Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ramendra Pati Pandey | -- | 2680 | 2023-02-08 08:55:44 | | | |
| 2 | Rita Xu | Meta information modification | 2680 | 2023-02-08 09:27:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Konyak, B.M.; Sharma, M.; Kharia, S.; Pandey, R.P.; Chang, C. Omicron Variant. Encyclopedia. Available online: https://encyclopedia.pub/entry/40970 (accessed on 08 February 2026).
Konyak BM, Sharma M, Kharia S, Pandey RP, Chang C. Omicron Variant. Encyclopedia. Available at: https://encyclopedia.pub/entry/40970. Accessed February 08, 2026.
Konyak, Beyau M., Mohan Sharma, Shabnam Kharia, Ramendra Pati Pandey, Chung-Ming Chang. "Omicron Variant" Encyclopedia, https://encyclopedia.pub/entry/40970 (accessed February 08, 2026).
Konyak, B.M., Sharma, M., Kharia, S., Pandey, R.P., & Chang, C. (2023, February 08). Omicron Variant. In Encyclopedia. https://encyclopedia.pub/entry/40970
Konyak, Beyau M., et al. "Omicron Variant." Encyclopedia. Web. 08 February, 2023.
Copy Citation
With the ongoing COVID-19 pandemic, the emergence of the novel Omicron variant in November 2021 has created chaos around the world.
emergence of Omicron and its mechanism
mutation and sub-lineages
monoclonal antibodies
1. Introduction
The emergence of this ongoing pandemic coronavirus has devastated the world. It has been almost two years since COVID-19 was detected in Wuhan, China. As of 12 August 2022, more than 6 million deaths and 585 million confirmed cases have been reported worldwide, making it one of the deadliest viruses in history [1]. This virus has undergone several recombination and mutations over the past years, leading to the emergence of a vast array of new variants of concern (VOC) and variants of interest (VOI), resulting in a high level of global health alerts and panic.
While the world has been tackling the Delta variant, the emergence of the novel Omicron variant in December 2021 took the world by storm [2]. The first SARS-CoV-2 variants to emerge were Alpha, Beta, Gamma, and Delta, and the Omicron (B.1.1.529) variant, classified as VOC by WHO [3] on 26 November 2021, has become the dominant one in many countries since January 2021 (Table 1). The novel Omicron variant has a larger number of mutations as compared to other VOCs. This has led to a higher rate of infections, higher transmissions, and immune escape from COVID-19 vaccines, resulting in rapid spread worldwide in a shorter period. As per the WHO, this novel variant also enhances immune or diagnostic evasion, causes less severe disease, and shows enhanced transmissibility. In addition, it poses a detrimental impact on epidemiology and shows increased variability in clinical presentation. The four most common symptoms of an Omicron infection, according to the US Centers for Disease Control and Prevention, are cough, fatigue, congestion, and a runny nose [4]. Although the Omicron variant seems more contagious than previous variants, as per the data available, early findings suggest lower rates of hospitalization for Omicron-positive patients, as compared to the Delta variant. However, the Omicron variant should not be taken lightly. Precautionary measures should be taken such as avoiding crowded spaces, maintaining social distances, and wearing masks to prevent the spread of infection [5]. The SARS-CoV-2 Interagency Group (SIG) evaluates and classifies the novel variant as a VOC, based on the cases detected in various countries, its travel history, rate of transmission, mutations in the spike protein, and as per data available for other variants. It is also evaluated based on a reduction in the effectiveness of vaccines and monoclonal antibody treatments. The SIG tracks and evaluates the emergence of new viruses and actively monitors the potential impact of vaccines, therapeutics, and diagnostics against SARS-CoV-2 [6].
Table 1. WHO-designated SARS-CoV-2 variants of concern (VOCs).
| WHO Designation | Country First Origin | Pango Lineages | Variant Prevalence Countries as of 11 February 2022 | GISAID | Next Strain | Mutation | Additional Amino Acid Changes Monitored |
|---|---|---|---|---|---|---|---|
| Alpha (18 December 2020) | United Kingdom, September-2020 |
9 Sub-lineages: B.1.1.7, Q.1, Q.4Q.5, Q.8, Q.7, Q.2, Q.6, Q.3. | United Kingdom (262,616) | GRY, GR/501Y.V1 | 20I/501Y.V1, 20B/501Y.V1 | 22 mutations (9 mutation spike protein, with deletion:del69/70, del144/144) |
+S:484K +S:452R |
| Beta (18 December 2020) | South Africa, May-2020 |
B.1.351, B.1.351.3, B.1.351.2, B.1.351.5, B.1.351.1 | South Africa (6885) | GH/501Y.V2 | 20H/501Y.V2 | 18 mutations (8 mutation at spike protein, with deletion:del241/243) | +S:L18F |
| Gamma (11 January 2021) | Brazil, November 2020. | 23 Pango lineages currently associated with the Gamma variant. | Brazil (47,475) | GR/501Y.V3 | 20J/501Y.V3 | 23 mutations (12 mutation at spike protein) | +S:681H |
| Delta (11 May 2021) | India, Oct-2020 |
216 Pango lineages currently associated with the Delta variant | India (69,457) | G/452R.V3 | 21A/S:478K | 29 mutations (8 mutation at spike protein, with deletion:del157/158) | +S:417N +S:484K |
| Omicron (26 November 2021) | South Africa, November 2021 | BA.1, BA.1.1, BA.2BA.3 | South Africa (4930) | GR/484A | 21K, 21L, 21M. | ∼50 mutations (30 mutations at spike protein, with deletion: del69/70, del143/145, and del212/212.) | +S:R346K |
2. The Emergence of the Omicron (B.1.1.529) Variant and Its Sub-Lineage
South Africa was the first country to detect the novel variant of SARS-CoV-2 from specimens collected on 11 November and 14 November 2021. The first Omicron variant was reported to WHO by the South African Department of Health on 24 November 2021 [6]. On 25 November 2021, the United Kingdom Health Security Agency designated the B.1.1.529 Omicron variant as a Variant Under Monitoring (VUI-21-NOV-01) and, just one day later, it was designated as a VOC by the WHO; the variant was later named Omicron [4][7]. The rapid spread of this variant in South Africa and surrounding countries alarmed the WHO. This could be due to the large number of mutations on the spike protein in the Omicron variant as compared to other VOCs. However, Omicron has been reported to be less severe than the previous variants [8][9]. Three days after the announcement of Omicron on 29 November 2021, by the WHO, this novel variant was detected in Australia, Belgium, Canada, the Czech Republic, Denmark, France, Germany, Italy, Netherlands, and the United Kingdom [10]. As of 25 December 2021, Omicron had been reported in 108 countries, with nearly 150,000 confirmed cases and 26 deaths, creating an alarming situation worldwide [11]. The emergence of the Omicron variant in South Africa has also revealed a vaccination gap between the richest and the poorest. Whilst an average of 60% of the population of Europeans has been vaccinated, it is appalling to see that only 5–10% of the African population has been vaccinated [12]. This could be one of the major factors contributing to the emergence of VOCs. Japan also reported the first two cases of the novel Omicron variant at the end of November 2021 among international travelers returning to the country with the undetected Omicron variant [13]. India reported the first two cases of the Omicron variant on 3rd December 2021 among international travelers [14]. In Germany, two suspected cases of the new Omicron variant were first reported on 27 November 2021 and, as the New Year began, this novel Omicron became one of the most dominant variants in Germany, despite protective measures at the initial stage in January 2022 [15][16]. This new variant has been spreading worldwide and has been reported in 180 countries as of 4 February 2022. As of 11 February 2022, the UK reported a maximum number of Omicron cases (422,993) followed by the USA (359,236), Denmark (69,789), Germany (48,866), Canada (30,687), and France (27,487), while an increasing number of cases can be seen on a daily basis in many other countries [16]. Jansen et al. [17] reported that the Omicron variant has a shorter incubation period than the previous variants, with similar clinical symptoms. Preliminary data conducted by the National Institute of Infectious Diseases shows a high viral load three to six days after the onset of symptoms [18].
As the Omicron variant continues to evolve and mutate, new sequences (sub-lineages) of the Omicron variant were reported by the WHO in December 2021, namely: BA.1, BA.2, and BA.3 [19]. Over the past two months, the BA.1 variant has escalated globally. However, in the last weeks of January 2022, the BA.2 sub-lineage had increased internationally and had already become dominant in Denmark. As of 11 February 2022, BA.2 had been detected in at least 70 countries and 44 U.S. states, according to the data uploaded in GISAID, and a total number of 55,404 sequences in the BA.2 lineage have been detected since the lineage was identified [20][21]. A preprint uploaded in medRxix [22], concluded that Omicron BA.2 is more dominant and transmissible than the parent Omicron variant (BA.1). It possesses immune-escaped advantages among vaccinated individuals; however, it does not increase its transmissibility to vaccinated individuals with breakthrough infections. India is another country where BA.2 is rapidly replacing the Delta and Omicron BA.1 variants, as per the report [23]. Data uploaded on GISAID suggest that the BA.2 variant is increasing rapidly in proportion to the original BA.1 variant, thus dominating the parental variant based on the sequence frequency [24].
3. Mechanism of Omicron Variant
The Omicron variant has the highest number of mutations observed so far, as compared to the other VOCs such as Alpha, Beta, Gamma, and Delta. This variant contains more than 30 mutations in the spike protein, which binds to the ACE2 receptor in the human cell for invasion, and out of which, 15 modifications are observed in a Receptor Binding Domain (RBD), an area strongly associated with humoral immune evasion [25], thus, increasing its transmissibility and thereby causing the possibility of evading vaccine-induced antibodies [26]. Researchers from China constructed and studied the binding affinity between the complex structure of the human ACE2 protein and the RBD of the Omicron variant spike protein (S-protein) using atomistic molecular dynamics simulations. They observed that mutations in the RBD of the SARS-CoV-2 Omicron variant resulted in a higher binding affinity to the human ACE2 protein [27].
There are two distinct mechanisms for the entry of SARS-CoV-2 into a human cell: cell-surface entry and endosomal entry, where TMPRSS2-mediated S protein activation takes place at the plasma membrane and cathepsin-mediated activation takes place in the endolysosome at the cytoplasmic region, respectively [28]. A researcher from Glasgow University in the UK demonstrated the replication process of different SARS-CoV-2 variants and their mechanisms in entering the human cell. An HIV-pseudotype was evaluated for entry assay. The study reveals that Omicron, like pangolin CoV, uses the endocytosis pathway to enter human cells and is independent of the TMPRSS2 present on the cell surface. This study was further supported by using drug inhibitors targeting either the TMPRSS2 (Camostat) or the cathepsins (E64d). As compared to another variant spike, the Omicron spike shows a reduced syncytium formation, thereby causing fewer lung infections [29].
4. Mutations in Omicron Variant
In November 2021, Omicron (B.1.1.529) emerged as a VOC. Wahid et al. [26] had earlier reported the triplet mutation (K417N + E484K + N501Y) in the Beta (B.1.351) and Gamma (P.1) variants, which were highly transmissible, leading to greater COVID-19 hospitalizations, ICU admissions, and even deaths. Such mutations have increased the immune escape potential leading to a decrease in the neutralization of antibodies (such as those found in the Pfizer and Moderna vaccines). The Omicron variant contains overall 50 mutations, with 30 mutations [25] on the spike protein, which include: A67V, del69/70, T95I, G142D, del143/145, N211I, del212/212, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F, of which 15 modifications are located at the RBD region [30]. Cecylia et al. [27] conducted atomistic molecular dynamics simulations to study the binding interactions between the human ACE2 protein and the RBD of the Omicron spike protein (S-protein). The analysis shows that the Omicron RBD binds more strongly to the human ACE2 protein than does the original Wuhan strain. However, the SARS-CoV-2 Omicron RBD shows weaker binding affinity than the Delta variant [31]. Most COVID-19 strains contain at least one change from the original Wuhan sequence, D614G, altering the virus’s ability to escape the immune response. The N501Y mutation is present in all VOCs, except the Delta variant [32]. Interestingly, Omicron’s spike protein mutations, such as D614G, N501Y, and K417N, are found in some other VOCs, thus making the virus more infectious—a prospect that is very concerning.
Similarly, the H655Y, N679K, and P681H mutations in the Omicron spike protein, also found in the Alpha and Delta variants, could increase the transmission of the virus [6][33][34]. It has the deletion at spike protein (Δ69–70), position 69–70, similar to the Alpha and Eta variants, that leads to the S-gene dropout or S-gene target failure. This phenomenon may provide a useful proxy to measure the prevalence of the Omicron variant. The rapid transmission of the Omicron variant across the globe could be due to the presence of these larger numbers of mutations in the spike protein, unlike the Delta variant, and thus evading immune response [33]. However, this phenomenon does not apply to the BA.2 sub-lineage of the Omicron variant, as it does not contain the deletion at S: 69–70 and is S-gene target positive (SGTP) on PCR diagnostic assays [35]. The comparison and mutual sharing of the spike protein mutations of the Omicron sub-lineages, BA.1, BA.2, and BA.3, are represented by a Venn diagram, as shown in Figure 1.
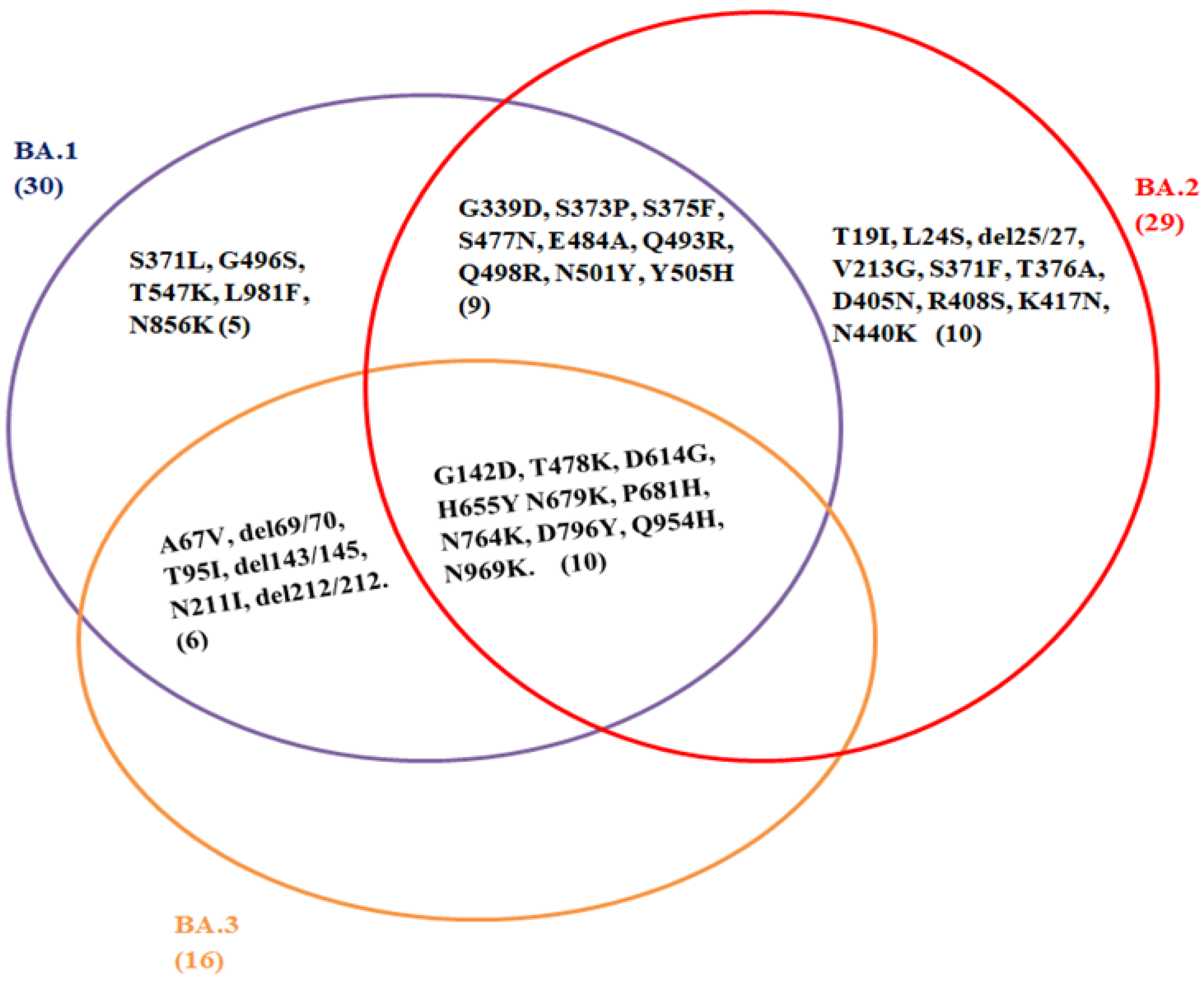
Figure 1. Comparison of spike protein mutations of Omicron sub-lineage: BA.1, BA.2, and BA.3. (GISAID: https://www.epicov.org/epi3/frontend#58aca) (Accessed on 10 February 2022). This Venn diagram represents the common mutation present among the sub-linages of Omicron variant; G142D, T478K, D614G, H655Y N679K, P681H, N764K, D796Y, Q954H, N969K. B.A.1 (30); A67V, del69/70, T95I, G142D, del143/145, N211I, del212/212, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F. B.A.2 (29); T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K. B.A.3 (16); A67V, del69/70, T95I, G142D, del143/145, N211I, del212/212, T478K, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K.
The combination of N501Y + Q498R may increase the binding affinity to ACE2, while other substitutions may lead to a decrease in the binding affinity to the ACE2 in the Omicron spike protein. Kinases’, such as PI3K/AKT, signaling are essential in SARS-CoV-2 entry. Based on molecular docking, Bexultan et al. [36] analyzed the interaction between the potential kinases and the N501Y mutation and observed that the N501Y mutation did not enhance binding to epidermal growth factor receptors (EGFR) due to the mutations. The N501Y mutation-containing lineages might become more infectious since several kinases are elevated in cancer patients. Therefore, additional care for cancer management should be taken into consideration. Mannar et al. [37] analyzed a cryo-electron microscopy structure between the spike proteins of the Omicron variant in complex with human ACE2. The structure depicts two additional new salt bridges and hydrogen bonds formed by the mutated residues R493, S496, and R498 in the RBD with the ACE2. This enhances other mutations, such as K417N, which is known to reduce the ACE2 binding affinity to the spike protein. These strong interactions at the ACE2 interface may contribute to the rapid spread of the Omicron variant [38]. Zhuoming et al. [39] reported that E484K could escape neutralization by convalescent sera, while S477N was resistant to neutralization by multiple mAbs, thus needing further investigation. The increase in binding affinity to the ACE2 receptor by N501Y could aid in an increase in transmission. The combination of N501Y and Q498R may also increase the binding affinity even more; however, other substitutions in the Omicron spike protein are expected to decrease binding to the ACE2. A cluster of mutations (such as H655Y, N679K, P681H) present at the S1–S2 furin cleavage site in the Omicron variant may increase spike cleavage and could aid in transmission. The N679K present at the furin cleavage site adds to the polybasic nature and is associated with an increase in transmission. The P681H mutation (also found in Alpha) enhances spike cleavage, and could also aid in transmission. At this position, an alternate mutation (P681R) is found in Delta [40]. In India, Surendra et al. [41] found a major mutation at the P681 position with an R (P681R) similar to the Delta variant (B.167.2) of about 9.71% instead of the H (P681H) mutation. This variant is of considerable public concern, as it is increasing at a high rate in many countries, including the U.S., due to its increased transmissibility and immune evasion. In the nucleocapsid (N) protein region, the Omicron variant also possesses two additional mutations such as R203K and G204R (found in ancestral mutation) that enhance sub-genomic RNA expression and viral RNA binding with key host proteins, thus increasing the viral load [42][43]. Phylogenetic analysis, based on the prevalence of high numbers of mutations, revealed that the Omicron variant is closely related to the Gamma (P.1) variant [44], and may possess similar characteristics at the molecular level [45].
References
- World Health Organization (WHO). WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. 2022. Available online: https://covid19.who.int/ (accessed on 14 February 2022).
- Rahmani, S.; Rezaei, N. Omicron (B.1.1.529) variant: Development, dissemination, and dominance. J. Med. Virol. 2021, 94, 1787–1788.
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants/ (accessed on 20 February 2022).
- World Health Organization (WHO). Classification of Omicron (B.1.1.529): SARSCoV-2 Variant of Concern. 2021. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 5 January 2022).
- UNICEF. What We Know about the Omicron Variant. Available online: https://www.unicef.org/coronavirus/what-we-know-about-omicron-variant (accessed on 16 February 2022).
- CDC. Science Brief: Omicron (B.1.1.529) Variant (Updated 2 December 2021). Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html (accessed on 16 February 2022).
- UK Health Security Agency. SARS-CoV-2 Variants of Concern and Variants under Investigation. 26 November 2021. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1036501/Technical_Briefing_29_published_26_November_2021.pdf (accessed on 14 February 2022).
- Harvard Medical School. Coronavirus Resource Center—Harvard Health. Harvard Health Publishing. Lab Studies, Animal Studies, and Epidemiological Data All Indicate That Omicron May Cause Less Severe Disease Than Previous Variants. Available online: https://www.health.harvard.edu/diseases-and-conditions/coronavirus-resource-center (accessed on 11 January 2022).
- Leonhardt, D. Omicron Is Milder. The New York Times, 5 January 2022. Available online: https://www.nytimes.com/2022/01/05/briefing/omicron-risk-milder-pandemic.html(accessed on 7 January 2022).
- Petersen, E.; Ntoumi, F.; Hui, D.S.; Abubakar, A.; Kramer, L.D.; Obiero, C.; Tambyah, P.A.; Blumberg, L.; Yapi, R.; Al-Abri, S.; et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529)—Highlights Africa’s research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2022, 114, 268–272.
- Mohapatra, R.K.; Sarangi, A.K.; Kandi, V.; Azam, M.; Tiwari, R.; Dhama, K. Omicron (B.1.1.529 variant of SARS-CoV-2); An emerging threat: Current global scenario. J. Med. Virol. 2021, 94, 1780–1783.
- WHO. Less than 10% of African Countries to Hit Key COVID-19 Vaccination Goal. 2021. Available online: https://www.afro.who.int/news/less-10-african-countries-hit-key-covid-19-vaccination-goal (accessed on 28 November 2021).
- Maruki, T.; Iwamoto, N.; Kanda, K.; Okumura, N.; Yamada, G.; Ishikane, M.; Ujiie, M.; Saito, M.; Fujimoto, T.; Kageyama, T.; et al. Two cases of breakthrough SARS-CoV-2 infections caused by the Omicron variant (B.1.1.529 lineage) in international travelers to Japan. Clin. Infect. Dis. 2022, 75, e354–e356.
- First Omicron Cases Detected in India. Available online: https://thehill.com/policy/international/583965-first-omicron-cases-detected-in-india (accessed on 6 January 2022).
- Germany Reports First Two Cases of Omicron Variant. Available online: https://www.thejournal.ie/germany-omicron-coronavirus-covid-19-bavaria-5613843-Nov2021/ (accessed on 6 January 2022).
- GISAID. Tracking of Variants. 2021. Available online: https://www.gisaid.org/hcov19-variants/ (accessed on 30 November 2021).
- Jansen, L.; Tegomoh, B.; Lange, K.; Showalter, K.; Figliomeni, J.; Abdalhamid, B.; Iwen, P.C.; Fauver, J.; Buss, B.; Donahue, M. Investigation of a SARS-CoV-2 B.1.1.529 (omicron) variant cluster—Nebraska, november-december 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1782–1784.
- National Institute of Infectious Diseases, Japan. Active Epidemiological Investigation on SARS-CoV-2 Infection Caused by Omicron Variant (Pango Lineage B.1.1.529 in Japan: Preliminary Report on Infectious Period. 2022. Available online: https://www.niid.go.jp/niid/en/2019-ncov-e/10884-covid19-66-en.html (accessed on 22 January 2022).
- UK Health Security Agency. SARS-CoV-2 Variants of Concern and Variants under Investigation in England. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050999/Technical-Briefing-35-28January2022.pdf (accessed on 6 February 2022).
- Washington Post. There’s a New Version of Omicron but So Far It Doesn’t Appear to Be More Dangerous. Available online: https://www.stripes.com/covid/2022-01-25/new-omicron-version-mutation-BA-2-4405690.html (accessed on 25 January 2022).
- Outbreak.Info. BA.2 Lineage Report. Available online: https://outbreak.info/situation-reports?pango=BA.2 (accessed on 11 February 2022).
- Lyngse, F.P.; Kirkeby, C.; Denwood, M.; Christiansen, L.E.; Mølbak, K.; Møller, C.H.; Skov, R.; Krause, T.; Rasmussen, M.; Sieber, R.; et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: Evidence from Danish Households. medRxiv 2022.
- Omicron BA.2: What We Know about the Covid Sub-Variant. Available online: https://www.bbc.com/news/health-60233899 (accessed on 7 February 2022).
- GISAID. hCoV-19 Tracking of Variants. Available online: https://www.epicov.org/epi3/frontend#58aca (accessed on 10 February 2022).
- Kupferschmidt, K.; Vogel, G. How bad is Omicron? Some clues are emerging. Science 2021, 374, 1304–1305.
- Wahid, M.; Jawed, A.; Mandal, R.K.; Dailah, H.G.; Janahi, E.M.; Dhama, K.; Somvanshi, P.; Haque, S. Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5857–5864.
- Lupala, C.S.; Ye, Y.; Chen, H.; Su, X.-D.; Liu, H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022, 590, 34–41.
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20.
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; Logan, N.; De Lorenzo, G.; Furnon, W.; Scott, S.; Manali, M.; Szemiel, A.; et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv 2022, 23, 3–20.
- Saxena, S.K.; Kumar, S.; Ansari, S.; Paweska, J.T.; Maurya, V.K.; Tripathi, A.K.; Abdel-Moneim, A.S. Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 2021, 94, 1738–1744.
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8.
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19.
- Corum, J.; Zimmer, C. Tracking Omicron and Other Coronavirus Variants. 2021. Available online: https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html (accessed on 7 January 2022).
- Poudel, S.; Ishak, A.; Perez-Fernandez, J.; Garcia, E.; León-Figueroa, D.A.; Romaní, L.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts—What is known so far? Travel Med. Infect. Dis. 2022, 45, 102234.
- Science the Wire. A ‘Stealth’ Version of Omicron Could Challenge Surveillance Efforts. Available online: https://science.thewire.in/external-affairs/world/ba-2-sublineage-omicron-s-gene-target-failure-surveillance/ (accessed on 7 January 2022).
- Kazybay, B.; Ahmad, A.; Mu, C.; Mengdesh, D.; Xie, Y. Omicron N501Y mutation among SARS-CoV-2 lineages: Insilico analysis of potent binding to tyrosine kinase and hypothetical repurposed medicine. Travel Med. Infect. Dis. 2022, 45, 102242.
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764.
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375, 846–868.
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.-M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e4.
- Quarleri, J.; Galvan, V.; Delpino, M.V. Omicron variant of the SARS-CoV-2: A quest to define the consequences of its high mutational load. Geroscience 2022, 44, 53–56.
- Negi, S.S.; Schein, C.H.; Braun, W. Regional and temporal coordinated mutation patterns in SARS-CoV-2 spike protein revealed by a clustering and network analysis. Sci. Rep. 2022, 12, 1128.
- Leary, S.; Gaudieri, S.; Parker, M.D.; Chopra, A.; James, I.; Pakala, S.; Alves, E.; John, M.; Lindsey, B.B.; Keeley, A.J.; et al. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: Homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. Pathog. Immun. 2021, 6, 27–49.
- Mourier, T.; Shuaib, M.; Hala, S.; Mfarrej, S.; Alofi, F.; Naeem, R.; Alsomali, A.; Jorgensen, D.; Subudhi, A.K.; Ben, R.F.; et al. SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load. Nat. Commun. 2022, 13, 601.
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022, 126, 102779.
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 2021, 398, 2126–2128.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
868
Revisions:
2 times
(View History)
Update Date:
08 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




