Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Garima Singh | -- | 2640 | 2023-02-03 06:20:10 | | | |
| 2 | Sirius Huang | -6 word(s) | 2634 | 2023-02-03 06:25:47 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Singh, G. Metagenomic Approach and Lichen Molecules Linked to Genes. Encyclopedia. Available online: https://encyclopedia.pub/entry/40794 (accessed on 07 March 2026).
Singh G. Metagenomic Approach and Lichen Molecules Linked to Genes. Encyclopedia. Available at: https://encyclopedia.pub/entry/40794. Accessed March 07, 2026.
Singh, Garima. "Metagenomic Approach and Lichen Molecules Linked to Genes" Encyclopedia, https://encyclopedia.pub/entry/40794 (accessed March 07, 2026).
Singh, G. (2023, February 03). Metagenomic Approach and Lichen Molecules Linked to Genes. In Encyclopedia. https://encyclopedia.pub/entry/40794
Singh, Garima. "Metagenomic Approach and Lichen Molecules Linked to Genes." Encyclopedia. Web. 03 February, 2023.
Copy Citation
Lichen secondary metabolites have tremendous pharmaceutical and industrial potential. Although more than 1000 metabolites have been reported from lichens, less than 10 have been linked to the genes coding them. Biosynthetic research focuses strongly on linking molecules to genes as this is fundamental to adapting the molecule for industrial application. Metagenomic-based gene discovery, which bypasses the challenges associated with culturing an organism, is a promising way forward to link secondary metabolites to genes in non-model, difficult-to-culture organisms.
biosynthetic gene clusters
secondary metabolites
atranorin
grayanic acid
gyrophoric acid
lecanoric acid
physodic/olivetoric acid
usnic acid
lichen metabolites
1. Metagenomic-Guided Linking of Molecules to Genes
The metagenomic-guided linking of metabolites to genes is a boon for unconventional, difficult-to-culture organisms, such as lichens. The structures of most of the secondary metabolites of lichen are already known. This serves as a baseline for predicting the biosynthetic machinery and gene/s required for the synthesis of a metabolite. The BGC-identifying algorithms, such as antiSMASH [1][2], predict the clusters present in the genomes and classify them according to the core gene that is present in them as an NRPS cluster, PKS cluster, terpene cluster, etc. AntiSMASH also compares the BGCs to the MiBiG [3], the database of BGCs characterized from plants, bacteria, and fungi, to identify those BGCs that are most structurally and functionally similar. This step already narrows down the candidate gene/cluster search substantially. The estimated BGCs can be further grouped into sets of structurally similar BGCs by clustering programs, such as BiG-SCAPE [4] and BiG-FAM [5], to exclude the structurally and functionally divergent BGCs. The phylogenetic clustering of the genes provides further evidence of the function of PKS [6][7][8][9]. In most cases (at least for the studies linking lichen metabolites to genes), this narrows down the search to a single, most suitable candidate PKS.
Next, the presence of suitable accessory genes (e.g., oxidases, methyltransferases) further ascertains the function of the cluster and authenticates if the cluster is equipped to code for a particular metabolite. The presence of an expected accessory gene in the cluster, for instance, an OMT in an O-methylated product and a CYP450 in a molecule involving an oxidation step for its synthesis, brings further evidence that the cluster most probably codes for the molecule in question.
2. Lichen Metabolites Linked to Genes
So far, the following lichen metabolites have been linked to genes: lecanoric acid (an orcinol didepside, Figure 1A), atranorin (a ß-orcinol depside, Figure 1B), grayanic acid (an orcinol didepsidone, Figure 1C), usnic acid (dibezofurane derivative, Figure 1D), gyrophoric acid (an orcinol tridepside), olivetoric acid (an orcinol didepside, Figure 1A), and physodic acid (an orcinol didepsidone, Figure 1C). Of these, the link has been experimentally validated for only two metabolites (Figure 2A,B), whereas, for the others, the inference has been based on genomic, phylogenetic and molecular data (Figure 2C–G).
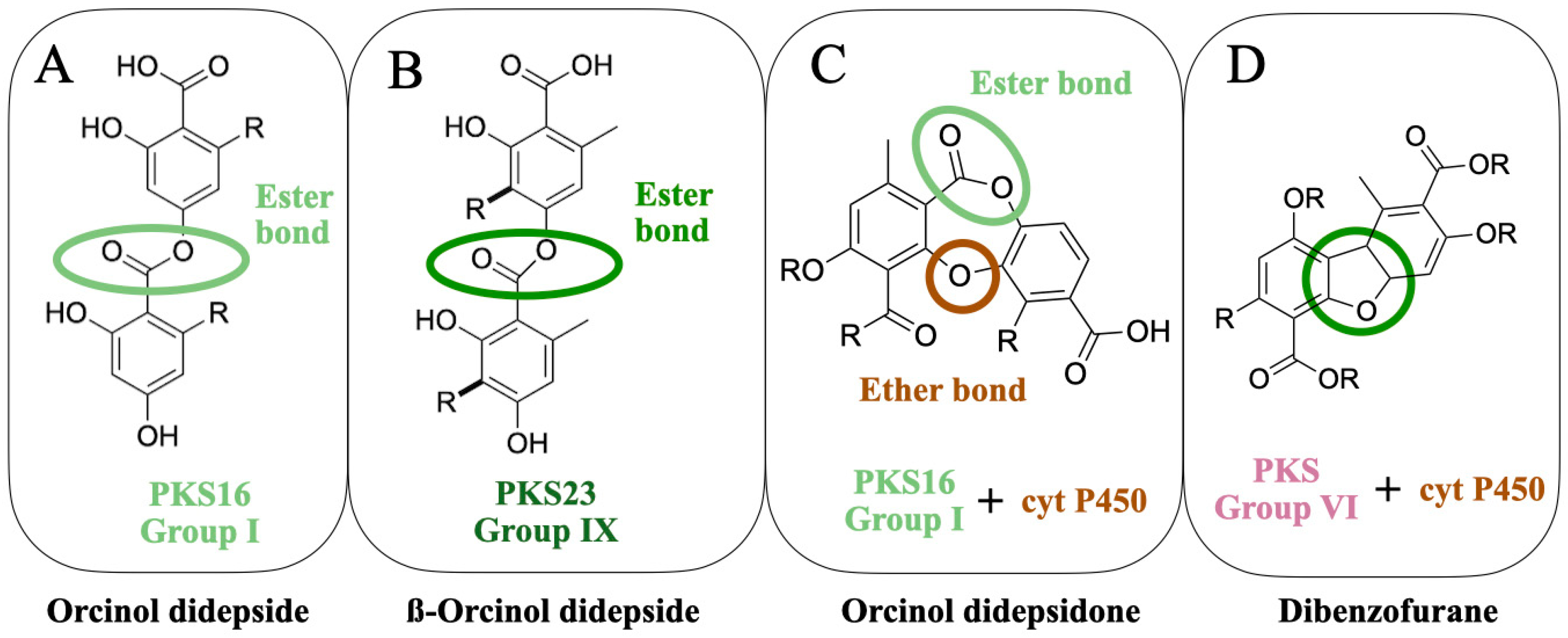
Figure 1. The molecular structures of four classes of lichen compounds linked to the genes. The colored circles denote the characteristic bond of a compound-ester bond present in a depside (A), the ester bond of a ß-orcinol depside (B), the ester and ether bond present in a depsidone (C), and the characteristic dibenzofurane bond (D).
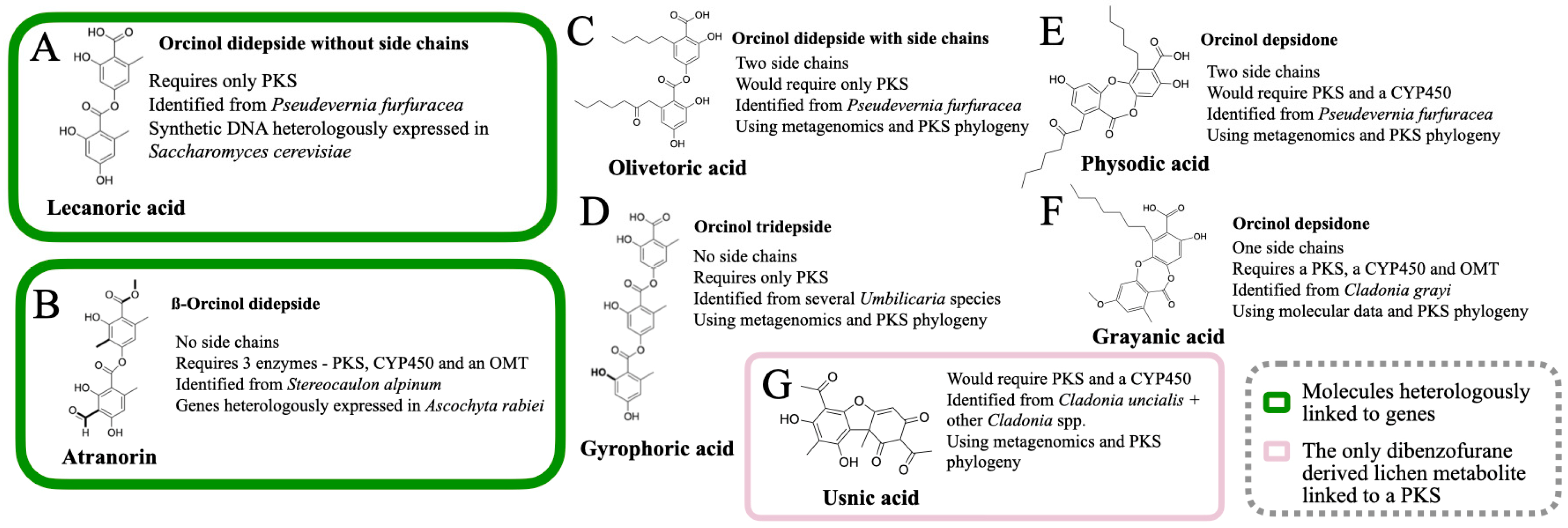
Figure 2. The lichen compounds that are linked to a PKS. The compounds highlighted in green are the ones in which the link between the molecule and the PKS was also verified by heterologous expression. (A) lecanoric acid, (B) atranorin, (C) olivetoric acid (D) gyrophoric acid, (E) physodic acid, (F) grayanic acid and (G) usnic acid.
2.1. Grayanic Acid
Grayanic acid (C21H22O7) is an orcinol depsidone (Figure 1C and Figure 2F). It is a relatively rare metabolite reported only from Neophyllis melacarpa and Cladonia grayi. Grayanic acid PKS was the first lichen PKS to be linked to the biosynthesis of a secondary metabolite [10].
The grayanic acid cluster was identified from Cladonia grayi in 2011 by integrating genome mining, phylogenetics, and other molecular biology approaches (Figure 2F), i.e., degenerate polymerase chain reaction (PCR) and the expression data analyses of single-spore isolates induced to produce grayanic acid [10]. The candidate cluster was singled out by comparing mRNA and grayanic acid induction profiles and by comparing the accumulation of grayanic acid to the induction of PKS. The expression of only one PKS is correlated to grayanic acid accumulation. The corresponding cluster has a cytochrome P450 monooxygenase (CYP450) and an O-methyltransferase gene, which are essential for the synthesis of grayanic acid (Figure 1C). This further validated the candidate PKS as the most likely grayanic acid PKS.
2.2. Atranorin
Atranorin (C19H18O8) is a ß-orcinol depside (Figure 1B and Figure 2B) with a broad-spectrum pharmacological property, e.g., analgesic, anti-inflammatory, antibacterial, antifungal, cytotoxic, antioxidant, and antiviral capabilities. This metabolite has been reported from several lichens, such as Bulbothrix spp., Cladonia spp., Evernia prunastri, Hypogymnia, Imshaugia, Letharia, Parmelia, Physia spp., Pseudevernia furfuracea, and Umbilicaria spp. Atranorin PKS has been so far identified from Bacidea rubella [11], Pseudevernia furfuracea [7][12], Cladonia rangiferina, Evernia prunastri, Letharia columbiana, Letharia lupina, Parmelia spp., and Stereocaulon alpinum [7].
Atranorin is one of the two lichen secondary metabolites to be successfully expressed heterologously [7]. The corresponding PKSs were identified for the first time in 2020 by integrating genome mining, BGC clustering, and the phylogeny approach, followed by experimental confirmation via heterologous expression [7]. The authors generated a NRPKS phylogeny using seven taxa—six Cladonia spp. and Stereocaulon alpinum. They used gene network analysis to group BGCs into gene cluster families and found that only one family was shared by the two atranorin producers, C. rangiferina and S. alpinum. These PKSs formed an additional supported clade in the PKS phylogeny, which is specific to the ß-orcinol compounds. This discovery led to the addition of an novel group, group IX, to the NRPKS phylogeny, in order to accommodate PKS23, the atranorin PKS.
The authors then identified five other atranorin BGCs using the CORASON pipeline [4] on the OMT sequence from C. rangiferina. Four genes, i.e., atranorin PKS, CYP450, OMT, and a transporter, were syntenic among the atranorin-producing species: atr1 (PKS23), atr2, atr3, and atr4. These four genes are conserved among the atranorin producing taxa used in above-mentioned study. It is worth noting that the atranorin cluster from both C. rangiferina and S. alpinum contains 14 genes, but only three were sufficient to produce the compound in a heterologous system.
The PKSs of group IX (PKSs coding for methylated orsellinic acid derivatives) were considered possible candidates for atranorin synthesis. The corresponding cluster had two other tailoring enzymes required for atranorin synthesis, namely, a CYP450 and an O-methyltransferase (Figure 2B). The candidate PKS from Stereocaulon alpinum, along with CYP450 and O-methyltransferase, was heterologously expressed in the plant-pathogenic fungus Ascochyta rabiei. The knock-in strain expressing the three genes produced atranorin, thereby definitively linking the cluster to atranorin synthesis (Figure 2B).
The study by Kim et al. [7] is an important milestone in lichen biochemistry. Not only was it the first time that a lichen compound was produced by heterologous expression, it also involved expressing three different genes from a cluster. The authors used an unconventional host, a plant pathogenic fungus, instead of Escherichia coli, Aspergillus nidulans, or A. oryzae. It is noteworthy that previous attempts to heterologously express lichen PKS using conventional hosts had failed [13]. Although the genes were transcribed, translation was not successful and the target metabolite was not obtained in the knock-in host. Bertrand and Sorensen 2019 [13], for instance, inserted four PKSs from Cladonia uncialis into A. oryzae and found that all of them were transcribed but none translated. The successful transcription and translation of lichen PKSs and the requisite accessory genes in Ascochyta rabiei to heterologously express atranorin could pave the way forward for the validation of other lichen PKSs, a hurdle that has restricted lichen biosynthetic studies for many years.
2.3. Lecanoric Acid
Lecanoric acid (C16H14O7) is an orcinol didepside (Figure 1A and Figure 1A) reported from several lichens including, Parmelia sp., Parmotrema sp., Umbilicaria sp. as well as non-lichenized fungi as Aspergillus sp. etc. It is one of the simplest didepsides, with two phenolic rings, joined together by an ester bond; the phenolic rings lack any side chains. Lecanoric acid is the other lichen metabolite that is heterologously expressed and is directly linked to a secondary metabolite [14]. The host and the approach that was used in this study were, however, different from the one used by Kim et al. [7].
The authors identified a candidate NRPKS from the Pseudevernia furfuracea genome, published under the framework of another study [15]. They used the putative grayanic acid NRPKS as bait for this. The protein sequence was deduced from the candidate NRPKS from P. furfuracea and was reverse-translated to generate a DNA sequence with codons optimized for Saccharomyces cerevisiae to synthesize the intron-free gene (6345 bp). The synthetic, intron-free and codon-optimized PKS gene, driven by the S. cerevisiae glucose-regulated ADH2 promoter, was introduced into S. cerevisiae via an expression vector. Yeasts harboring an expression plasmid produced lecanoric acid, thereby directly linking this PKS to lecanoric acid synthesis.
This study by Kealey et al. [14], however, raises some questions: lecanoric acid (Figure 2A) is not produced by P. furfuracea in nature; instead, the orcinol compounds reported from P. furfuracea are olivetoric acid (a didepside, Figure 2C) and physodic acid (a didepsidone, Figure 2E). As there is only one PKS16 homolog (depside PKS) present in P. furfuracea, the “lecanoric acid” PKS is, thus, involved in the synthesis of olivetoric/physodic acid. The experimental justification for “how a putative olivetoric/physodic acid PKS produces lecanoric acid in yeast” is yet derived but it was proposed that this could be attributed to the lack of appropriate starter units in yeast [12]. This conundrum gave rise to an interesting hypothesis regarding polyketide synthesis, i.e., the starter unit is crucial to enhancing the diversity of secondary metabolites, as the same PKS may produce different compounds in the presence of different starter units [12]. In fact, the authors themselves tried to feed different starter units but they always obtained lecanoric acid.
The study by Kealey et al. [14] marks an important milestone in lichen biochemical studies, first because it unequivocally links an NRPKS, PKS16, to the synthesis of an orcinol depside (Figure 1A), confirming the role of these PKSs in the synthesis of orcinol compounds, and second, because it highlights the importance of the host and the starter units in the heterologous expression of lichen secondary metabolites.
3. Molecules That Are Bioinformatically Linked to Their Respective Genes/Clusters
3.1. Usnic Acid
Usnic acid (C18H16O7) is a dibenzofuran derivative (Figure 1D and Figure 2G). The usnic acid gene cluster is one of the best-studied lichen biosynthetic clusters, both genomically and experimentally, as well as across species. It is reported from several lichenized fungal genera, including Alectoria, Cladonia, Evernia, Lecanora, Ramalina, Usnea, and Usnochroma. The first most-likely gene cluster for usnic acid was identified from Cladonia uncialis in 2016 [16]. To date, this cluster has been identified from about 20 lichenized fungi, including C. rangiferina, C. metacorallifera, Lobaria pulmonaria, Nephromopsis pallescens [17], and Usnea spp. (Figure 2G). The putative usnic acid PKS belongs to group VI.
The first usnic acid gene cluster (from Cladonia uncialis) was identified via genome mining approach [16]. The authors narrowed down the most suitable candidate NRPKS from C. uncialis based on the biosynthetic requirements for usnic acid synthesis, i.e., a, NRPKS with a carbon methylation (cMT) domain, a terminal Claisen cyclase (CLC) domain, and an oxidative enzyme (mostly CYP450) for the dimerization of methylphloracetophenone to usnic acid. Only one candidate was found to fulfill the above-stated requirements and the corresponding genes were also transcriptionally active, suggesting that this may be the most likely usnic acid cluster.
Later, a phylogeny of NRPKSs from 46 lichenized fungi, including Cladonia uncialis, showed the presence of putative usnic acid PKS in all the producers and their absence in the non-producers [18]. Although the overall cluster composition varies, all producers have the usnic acid PKS and a CYP450 gene, as found in C. uncialis, building further evidence that the candidate usnic acid PKS that has been identified in C. uncialis was the one involved in usnic acid synthesis. It is to be noted that in all these taxa, usnic acid was a major metabolite and was identified using TLC or HPLC. Recently, usnic acid (and the corresponding cluster) was reported from C. rangiferina, formerly considered a non-producer, using LC-MS, a more sensitive detection technique [19]. This indicates that the presence of usnic acid might be more abundant in lichenized fungi than we currently know. Implementing more sensitive metabolite detection techniques will be essential to understand the taxonomic breadth and evolution of this gene cluster.
3.2. Olivetoric/Physodic Cluster
Olivetoric acid (C26H32O8) and physodic acid (C26H30O8) (Figure 2C,E) are potent cytotoxic, antimicrobial, and anti-oxidative agents. They are orcinol derivatives and comprise isostructural depside-depsidone pairs, i.e., they are structurally similar compounds except for the fact that olivetoric acid is an orcinol didepside (Figure 1A), whereas physodic acid is the an orcinol didepsidone, most probably derived from the oxidation of olivetoric acid by a CYP450 (Figure 1C). These two are reported to co-occur in the same species, often within the same sample but in different proportions—Pseudevernia furfuracea, Cetraria ciliaris, Ramalina leoidea, and Hypogymnia lugubris [12][20][21][22][23] The corresponding organisms are considered to be chemical races, based on the compounds present in them, but, often, both metabolites are reported to be present in the same sample.
The olivetoric/physodic cluster was recently identified from Pseudevenia furfuracea, based on the metagenomic and transcriptomic data [12]. The chemical race of the sample was first determined by HPLC, followed by the whole-genome sequencing of the races. Comparative genomics revealed only one PKS16 in both samples. The PKS16 from the two chemotypes was homologous, as expected, since the backbone molecule for both molecules is the same, except for the fact that there is an additional ether bond in physodic acid. Given that PKS16 catalyzes only ester bond synthesis and, therefore, depside formation (Figure 1A), it is quite likely that PKS produces the depside, olivetoric acid, in both chemotypes but in the physodic acid chemotype the CYP450 is active and catalyzes the synthesis of the depsidone.
The study by Singh et al. [12] shows that a cluster may be involved in the synthesis of more than one compound. This has been shown in non-lichenized fungi, but this was the first indication of the occurrence of such a phenomenon in lichenized fungi. In addition, the same PKS, when expressed heterologously in yeast, produced lecanoric acid, which is, again, a didepside, further demonstrating the promiscuity of this PKS. The same PKS synthesizes three different molecules, depending upon the starter unit (acetyl CoA, malonyl CoA, hexanoyl CoA, or octanoyl CoA) and the accessory enzymes that are activated (CYP450 is present upstream of the PKS when activated; it catalyzes the ether bond formation, leading to physodic acid synthesis).
3.3. Gyrophoric Cluster
Gyrophoric acid (C24H20O10) is a tridepside (Figure 2D) that is synthesized by several Umbilicaria species, Cryptothecia rubrocincta, Lecidea fuscoatra, Montanelia tominii, Parmotrema tinctorum, Punctelia borreri and Xanthoparmelia pokomyi etc. Usually, it is accompanied by several other structurally related molecules, such as umbilicaric acid, hiascic acid, and lecanoric compounds.
The most-likely gyrophoric acid cluster has been recently identified from nine Umbilicaria species [24]. Interestingly, although the species had different minor metabolites (U. deusta and U. grisea had umbilicaric acid, while U. sprodochroa had umbilicaric and lecanoric acid), only one copy of PKS16 was present in all Umbilicaria species included in the study by Singh et al. [24]. The corresponding PKS, PKS16, was highly homologous among all species. This study further indicates that PKS16 may be promiscuous and may synthesize a few structurally-related compounds. Another highlight of this study was that it was the first work identifying a tridepside PKS. This study showed that the domain structures of a tridepside PKS and a didepside PKS are the same, i.e., a KS-AT-ACP-ACP-TE, and that a third ACP domain might not be required for tridepside synthesis.
References
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35.
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346.
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: A Community-Driven Effort to Annotate Experimentally Validated Biosynthetic Gene Clusters. Nucleic Acids Res. 2022, 51, 603–610.
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16, 60–68.
- Kautsar, S.A.; Blin, K.; Shaw, S.; Weber, T.; Medema, M.H. BiG-FAM: The Biosynthetic Gene Cluster Families Database. Nucleic Acids Res. 2021, 49, D490–D497.
- Liu, L.; Zhang, Z.; Shao, C.-L.; Wang, J.-L.; Bai, H.; Wang, C.-Y. Bioinformatical Analysis of the Sequences, Structures and Functions of Fungal Polyketide Synthase Product Template Domains. Sci. Rep. 2015, 5, 10463.
- Kim, W.; Liu, R.; Woo, S.; Kang, K.B.; Park, H.; Yu, Y.H.; Ha, H.-H.; Oh, S.-Y.; Yang, J.H.; Kim, H.; et al. Linking a Gene Cluster to Atranorin, a Major Cortical Substance of Lichens, through Genetic Dereplication and Heterologous Expression. mBio 2021, 12, e0111121.
- Navarro-Muñoz, J.C.; Collemare, J. A Bioinformatics Workflow for Investigating Fungal Biosynthetic Gene Clusters. In Engineering Natural Product Biosynthesis: Methods and Protocols; Skellam, E., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 1–21. ISBN 978-1-07-162273-5.
- Kroken, S.; Glass, N.L.; Taylor, J.W.; Yoder, O.C.; Turgeon, B.G. Phylogenomic Analysis of Type I Polyketide Synthase Genes in Pathogenic and Saprobic Ascomycetes. Proc. Natl. Acad. Sci. USA 2003, 100, 15670–15675.
- Armaleo, D.; Sun, X.; Culberson, C. Insights from the First Putative Biosynthetic Gene Cluster for a Lichen Depside and Depsidone. Mycologia 2011, 103, 741–754.
- Gerasimova, J.V.; Beck, A.; Werth, S.; Resl, P. High Diversity of Type I Polyketide Genes in Bacidia rubella as Revealed by the Comparative Analysis of 23 Lichen Genomes. J. Fungi 2022, 8, 449.
- Singh, G.; Armaleo, D.; Dal Grande, F.; Schmitt, I. Depside and Depsidone Synthesis in Lichenized Fungi Comes into Focus through a Genome-Wide Comparison of the Olivetoric Acid and Physodic Acid Chemotypes of Pseudevernia furfuracea. Biomolecules 2021, 11, 1445.
- Bertrand, R.L.; Sorensen, J.L. Lost in Translation: Challenges with Heterologous Expression of Lichen Polyketide Synthases. ChemistrySelect 2019, 4, 6473–6483.
- Kealey, J.T.; Craig, J.P.; Barr, P.J. Identification of a Lichen Depside Polyketide Synthase Gene by Heterologous Expression in Saccharomyces cerevisiae. Metab. Eng. Commun. 2021, 13, e00172.
- Calchera, A.; Dal Grande, F.; Bode, H.B.; Schmitt, I. Biosynthetic Gene Content of the “Perfume Lichens” Evernia prunastri and Pseudevernia furfuracea. Molecules 2019, 24, 203.
- Abdel-Hameed, M.; Bertrand, R.L.; Piercey-Normore, M.D.; Sorensen, J.L. Putative Identification of the Usnic Acid Biosynthetic Gene Cluster by de Novo Whole-Genome Sequencing of a Lichen-Forming Fungus. Fungal Biol. 2016, 120, 306–316.
- Wang, Y.; Geng, C.; Yuan, X.; Hua, M.; Tian, F.; Li, C. Identification of a Putative Polyketide Synthase Gene Involved in Usnic Acid Biosynthesis in the Lichen Nephromopsis pallescens. PLoS ONE 2018, 13, e0199110.
- Pizarro, D.; Divakar, P.K.; Grewe, F.; Crespo, A.; Dal Grande, F.; Lumbsch, H.T. Genome-Wide Analysis of Biosynthetic Gene Cluster Reveals Correlated Gene Loss with Absence of Usnic Acid in Lichen-Forming Fungi. Genome Biol. Evol. 2020, 12, 1858–1868.
- Egbert, S.; Hoffman, J.R.; McMullin, R.T.; Lendemer, J.C.; Sorensen, J.L. Unraveling Usnic Acid: A Comparison of Biosynthetic Gene Clusters between Two Reindeer Lichen (Cladonia rangiferina and C. uncialis). Fungal Biol. 2022, 126, 697–706.
- Posner, B.; Feige, G.B. Studies on the Chemistry of the Lichen Genus Umbilicaria Hoffm. Z. Für Nat. C 1992, 47, 1–9.
- Walker, J.R.L.; Lintott, E.A. A Phytochemical Register of New Zealand Lichens. N. Z. J. Bot. 1997, 35, 369–384.
- Culberson, C.F. Joint Occurrence of a Lichen Depsidone and Its Probable Depside Precursor. Science 1964, 143, 255–256.
- Culberson, W.L.; Culberson, C.F.; Johnson, A. Pseudevernia Furfuracea-Olivetorina Relationships: Chemistry and Ecology. Mycologia 1977, 69, 604–614.
- Singh, G.; Calchera, A.; Merges, D.; Valim, H.; Otte, J.; Schmitt, I.; Dal Grande, F. A Candidate Gene Cluster for the Bioactive Natural Product Gyrophoric Acid in Lichen-Forming Fungi. Microbiol. Spectr. 2022, 10, e0010922.
More
Information
Subjects:
Genetics & Heredity
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
886
Revisions:
2 times
(View History)
Update Date:
03 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





