Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Li, P.; Zhang, S.; Wang, J.; Al-Shamiri, M.M.; Han, B.; Chen, Y.; Han, S.; Han, L. Secretion Systems of Acinetobacter baumannii. Encyclopedia. Available online: https://encyclopedia.pub/entry/40727 (accessed on 02 February 2026).
Li P, Zhang S, Wang J, Al-Shamiri MM, Han B, Chen Y, et al. Secretion Systems of Acinetobacter baumannii. Encyclopedia. Available at: https://encyclopedia.pub/entry/40727. Accessed February 02, 2026.
Li, Pu, Sirui Zhang, Jingdan Wang, Mona Mohamed Al-Shamiri, Bei Han, Yanjiong Chen, Shaoshan Han, Lei Han. "Secretion Systems of Acinetobacter baumannii" Encyclopedia, https://encyclopedia.pub/entry/40727 (accessed February 02, 2026).
Li, P., Zhang, S., Wang, J., Al-Shamiri, M.M., Han, B., Chen, Y., Han, S., & Han, L. (2023, February 01). Secretion Systems of Acinetobacter baumannii. In Encyclopedia. https://encyclopedia.pub/entry/40727
Li, Pu, et al. "Secretion Systems of Acinetobacter baumannii." Encyclopedia. Web. 01 February, 2023.
Copy Citation
Infections led by Acinetobacter baumannii strains are of great concern in healthcare environments due to the strong ability of the bacteria to spread through different apparatuses and develop drug resistance. Secretion systems have recently been demonstrated to be involved in the pathogenic process, and five types of secretion systems out of the known six from Gram-negative bacteria have been found in A. baumannii. They can promote the fitness and pathogenesis of the bacteria by releasing a variety of effectors. Additionally, antibiotic resistance is found to be related to some types of secretion systems.
Acinetobacter baumannii
secretion systems
pathogenicity
1. Introduction
Acinetobacter baumannii is a strictly aerobic, non-fermenting, Gram-negative coccobacillus with pili and capsule, but no flagella. It is ubiquitous in nature, and used to be considered to be of negligible significance due to its low virulence [1]. However, the rapidly increasing nosocomial infections and high mortality caused by A. baumannii, as well as its strong drug resistance, have raised people’s attention [2]. Taken together with Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Pseudomonas aeruginosa, Enterobacter, Acinetobacter baumannii has been enrolled as a member of ESKAPE by the Infectious Diseases Society of America (IDSA) in order to emphasize the importance of these pathogens in causing hospital infections and resisting the effects of a variety of antimicrobial drugs [3][4].
The high frequency of A. baumannii nosocomial infections is closely related to its strong environmental persistence. A. baumannii can survive in nutrient-limited and desiccation environments, and is capable of resisting disinfections [5]. Moreover, it is able to survive for long periods of time on both biotic and abiotic surfaces [6]. Based on these advantages, A. baumannii can be easily transmitted patient to patient by air, water, and contact with medical personnel’s hands and equipment, thus colonizing multiple sites and finally leading to a variety of infections, such as pneumonia, septicemia, urinary tract infections, meningitis, and skin and wound infections [7][8][9].
Antibiotic resistance is another key factor that contributes to A. baumannii infections and outbreaks. The increasing rate of infections caused by drug-resistant A. baumannii is a significant issue in hospitals all over the world [10]. The continued overuse and misuse of antibiotics enable A. baumannii to develop different types of resistance mechanisms, e.g., the acquisition of multiple antibiotic resistance genes to produce degradative enzymes, a decrease in bacterial membrane permeability, the alteration of antibiotic targets, the overexpression of efflux pumps, a change in metabolic status, and the formation of biofilms [11]. Therefore, this bacterium can escape the killing of antibiotics and conquer the stress conditions, further leading to infections. A. baumannii has an extraordinary genetic plasticity that results in a high capacity to acquire antimicrobial resistance traits [2], thus producing many multidrug-resistant (MDR), extensively drug-resistant (XDR), and even pan-drug-resistant (PDR) strains, representing a significant challenge for therapy in clinics.
Infections are also dependent on virulence factors. Various genes have been revealed to be involved in the pathogenic procedures of iron acquisition, nutrient uptake, adhesion, biofilm formation, invasion, hemolytic activity, and cytolytic activity [12][13]. Among them, protein secretion systems have received much attention. They can transport the virulence factors produced by bacteria into extracellular environments, meaning that the latter will manipulate the host’s defenses and facilitate pathogen infection [14][15]. Until recently, six secretion systems from Gram-negative bacteria have been revealed and studied; namely, type I secretion system (T1SS) to type VI secretion system (T6SS). Some of these have been characterized and reported to have specific roles in the pathophysiology of A. baumannii, whereas the gene and protein structures of some secretion systems in A. baumannii are still not clear and are being explored. Moreover, the association between secretion systems and drug resistance has been discovered in some bacteria, e.g., the T3SS in Pseudomonas aeruginosa correlates with a fluoroquinolone resistance phenotype, and the T4SS in many Gram-negative pathogens mediates antibiotic resistance via conjugation [16][17][18]. Meanwhile, the contribution of secretion systems to antibiotic resistance in A. baumannii is poorly understood.
2. Type I Secretion System (T1SS)
The T1SS is a highly conserved secretion system in pathogenic Gram-negative bacteria. However, it is less reported in A. baumannii. In 2017, the T1SS was first identified in the pathogenic Acinetobacter nosocomialis strain M2 upon bioinformatic analysis by Harding et al. [19]. Until now, only two reports have described the structure and function of the T1SS in Acinetobacter [19][20].
Gene and Structure
The locus that is homologous to the prototype T1SS of Escherichia coli containing the tolC, hlyB, and hlyD genes is found in the M2 chromosome, as well as in A. baumannii. In contrast to E. coli, these genes are found in three gene clusters, and are most likely in an operon, given that the open reading frame (ORF) for hlyB overlaps with both tolC and hlyD [19] (Figure 1a).
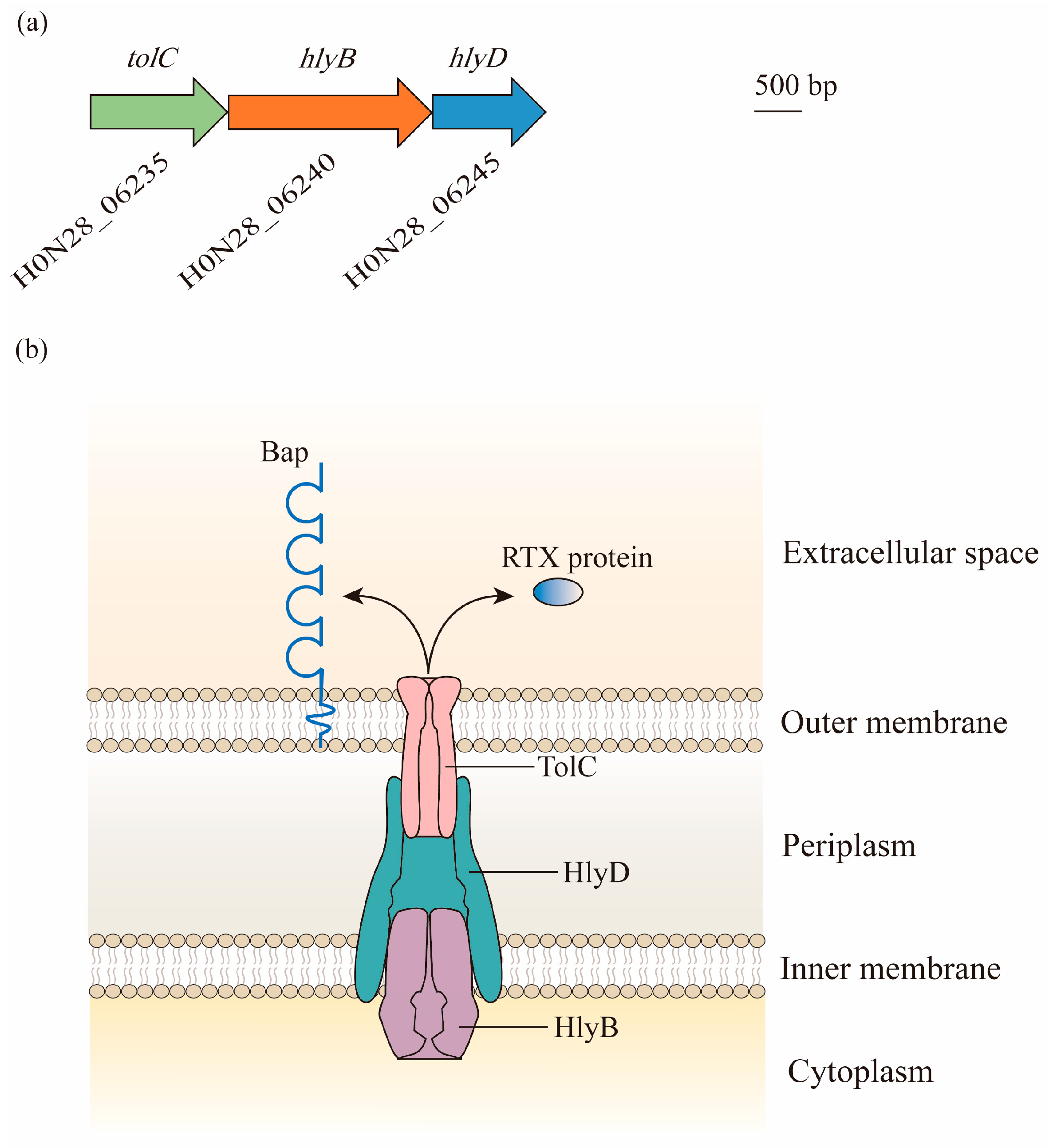
Figure 1. Composition and structure of the type I secretion system (T1SS) in A. baumannii: (a) Bioinformatic analysis has led to the identification of the T1SS in genomes of A. baumannii. Gene locus tags are cited from ATCC 17978. Genes predicted to encode proteins required for the biogenesis of the T1SS are found in three gene clusters, with hlyB overlapping with tolC and hlyD. (b) The three components of the T1SS act together to facilitate the secretion of effectors. TolC is a trimeric outer membrane protein with the α-helical barrel forming a tunnel through the periplasm, and it interacts with HlyD. HlyD has a large periplasmic domain linked by a single transmembrane helix, which anchors in the inner membrane. The energy required for the export of specific T1SS substrates is provided by HlyB, which is an ATP-binding protein. Two putative T1SS effectors, namely, Repeats-in-Toxin (RTX)-serralysin-like toxin and biofilm-associated protein (Bap), are involved in the formation and stability of biofilm.
This tolC-hlyB-hlyD gene cluster produces three proteins with high molecular weights of 130 kDa, 250 kDa, and 70 kDa. They form a secretion system with the elements of TolC, which is localized in the outer membrane, HlyB, which is anchored in the inner membrane as an ATP-binding cassette transporter, and HlyD as a periplasmic adaptor [19] (Figure 1b).
3. Type II Secretion System (T2SS)
The T2SS is a multiprotein secretion system that is widely distributed in Gram-negative bacteria, including enterotoxigenic Escherichia coli, Legionella pneumophila, Vibrio cholerae, Pseudomonas aeruginosa, and Klebsiella pneumoniae [21][22][23][24][25][26][27]. It was first reported in A. baumannii in 2014 and was subsequently shown to be active in ATCC 17978 by Johnson et al. [28][29]. Further, the T2SS is found in the majority of A. baumannii genomes.
Gene and Structure
In Acinetobacter spp., the T2SS is encoded by 12 essential genes, namely, gspC-M and pilD, and forms an apparatus spanning both the inner membrane and outer membrane [30][31] (Figure 2). In contrast to other Gram-negative pathogens, the core gsp genes are not organized in one or two operons, but are grouped into five distinct gene clusters scattered throughout the Acinetobacter genome [32] (Figure 2a).
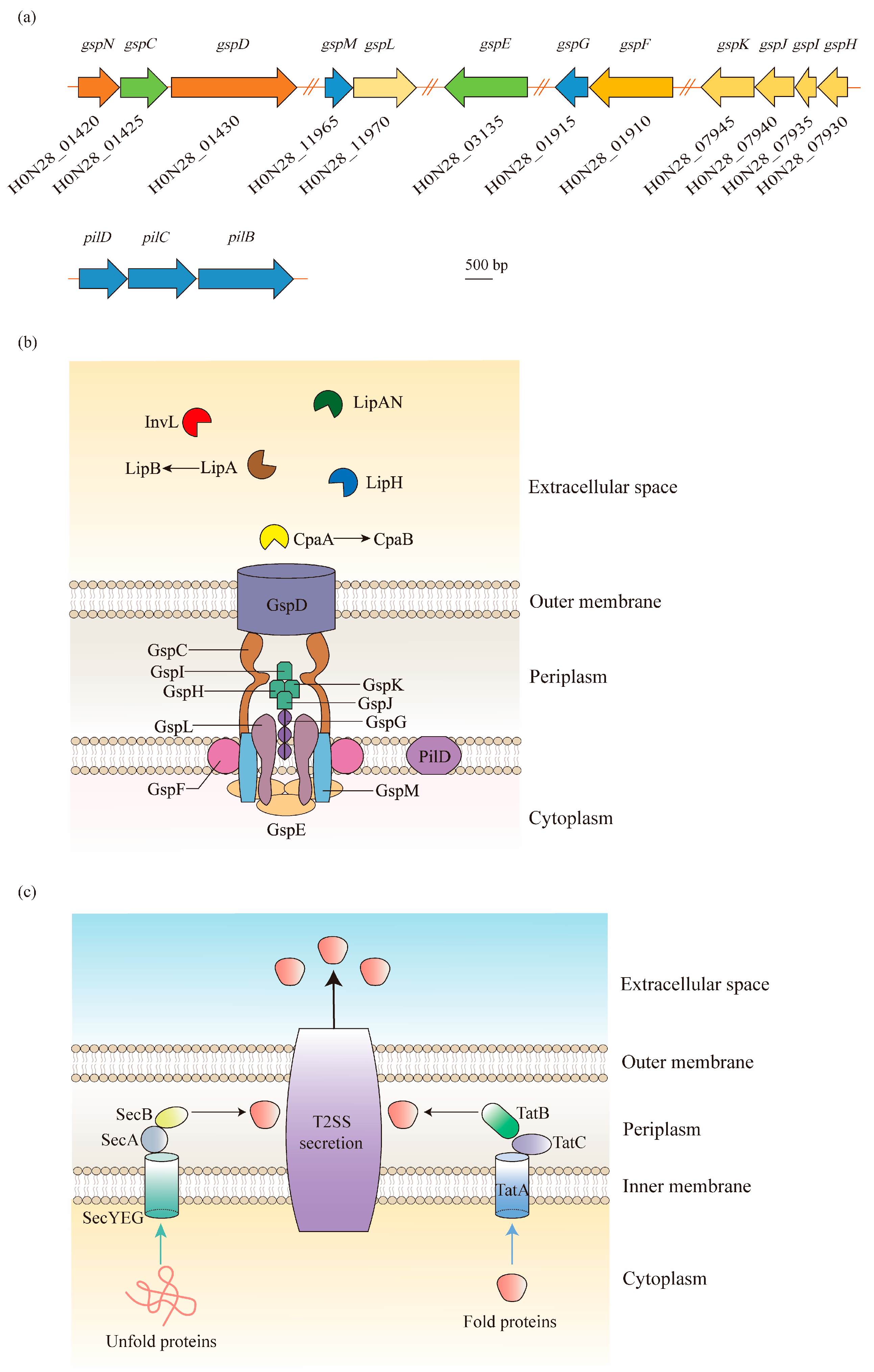
Figure 2. Type II secretion system (T2SS) structure of A. baumannii and its protein secretion mechanism: (a) As shown in the ATCC 17978 genome, the gsp genes required for the T2SS are located in five distant loci, and a single prepilin/pre-pseudopilin peptidase homolog is located in the pilBCD cluster. (b) The T2SS is composed of an outer membrane (OM) complex (GspD), a periplasmic pseudopilus (GspG, GspH, GspI, GspJ, and GspK), and an inner membrane (IM) platform (GspC, GspF, GspL, and GspM), which relates to the cytoplasmic ATPase GspE. In A. baumannii, the T2SS shares a processing protein, PilD, with type IV pili. The T2SS secretes a large number of effectors required for virulence, including the metallopeptidase CpaA (chaperone CpaB), the lipoyl synthases LipA (chaperone LipB), LipH, and LipAN, and a novel lipoprotein, InvL. (c) The T2SS-dependent proteins are first exported across the IM to the periplasm via the Sec or Tat pathways in A. baumannii. The Sec pathway primarily translocates unfolded proteins, relying on a hydrophobic signal sequence at the N-terminus. On the contrary, the Tat pathway, consisting of TatA, TatB, and TatC, primarily secretes folded proteins. Afterwards, the signal sequence is cleaved, followed by the folding of proteins. Finally, the folded proteins are expelled extracellularly through the OM channel.
In general, the T2SS consists of four parts: (1) an outer membrane (OM) complex; (2) a periplasmic pseudopilus; (3) an inner membrane (IM) complex called the assembly platform (AP); and (4) a cytoplasmic ATPase [33]. The OM complex is composed of GspD, which forms a secretin channel across the outer membrane to transport substrates from the periplasm to the extracellular milieu [34]. The IM platform is composed of GspC, GspF, GspL, and GspM, in which GspC is joined to the periplasmic domains of GspD, thereby connecting the IM platform with the OM complex. In between the OM and IM complexes, the periplasmic pseudopilus, a structure homologous to the type IV pilus, is attached to the IM platform with the composition of major pseudopilin GspG and minor pseudopilins GspH, GspI, GspJ, and GspK. Before the assembly of these subunits, PilD is involved in the cleavage and methylation procedure. Additionally, the cytoplasmic ATPase is formed by a hexamer protein, GspE, to provide ATP to the T2SS for the secretion of effector proteins [33][35] (Figure 2b).
4. Type IV Secretion System (T4SS)
T4SSs are multiprotein nanomachines, widespread in Gram-negative and Gram-positive bacteria, that deliver macromolecules, e.g., DNA and protein, to bacterial recipients or eukaryotic target cells [36]. They are generally divided into three groups; namely, type F and P (IVA), IVB, and GI systems [37][38]. However, T4SSs are less reported in A. baumannii. The information can be summarized from five studies, as discussed below. By using the high-density pyrosequencing method, the elements homologous to the Legionella/Coxiella T4S apparatus were first discovered in A. baumannii ATCC17978 [39]. Later, in a pathogenic isolate, ACICU, the plasmid pACICU2 was found harboring a complete tra locus, which encoded the conjugative apparatus and an F-type T4SS (based on the F-plasmid of Escherichia coli) [40]. However, the structure and function of the A. baumannii T4SS were not illustrated in these two studies. Furthermore, the plasmid replicase (rep) gene repAci6 from pACICU2 was found widely distributed in A. baumannii clinical strains, which carried the T4SS protein TraC coding gene [41][42]. Thus, repAci6 served as a candidate for screening the F-type T4SS, and the plasmid carried the genes required for the biogenesis of the T4SS, such as traC, traD, and traU, which were identified in clinical carbapenem-resistant A. baumannii (CRAB) isolates [43].
Gene and Structure
The F-type T4SS in A. baumannii contains a series of tra operon genes, including traA, traB, traC, traD, traE, traF, traG, traH, traI, traK, traL, traM, traN, traU, traV, and traW, as well as another two genes, trbC and finO. Through the alignment of seven F-like A. baumannii plasmids, it was observed that the core genes involved in pilus biosynthesis (traA, traB, traC, traF, traH, traK, traU, traV, traW, and trbC), nicking (traI), the initiation of transfer (traM and traD), mating aggregate stabilization (traN and traG), and regulation (finO) were highly conserved [43] (Figure 3a).
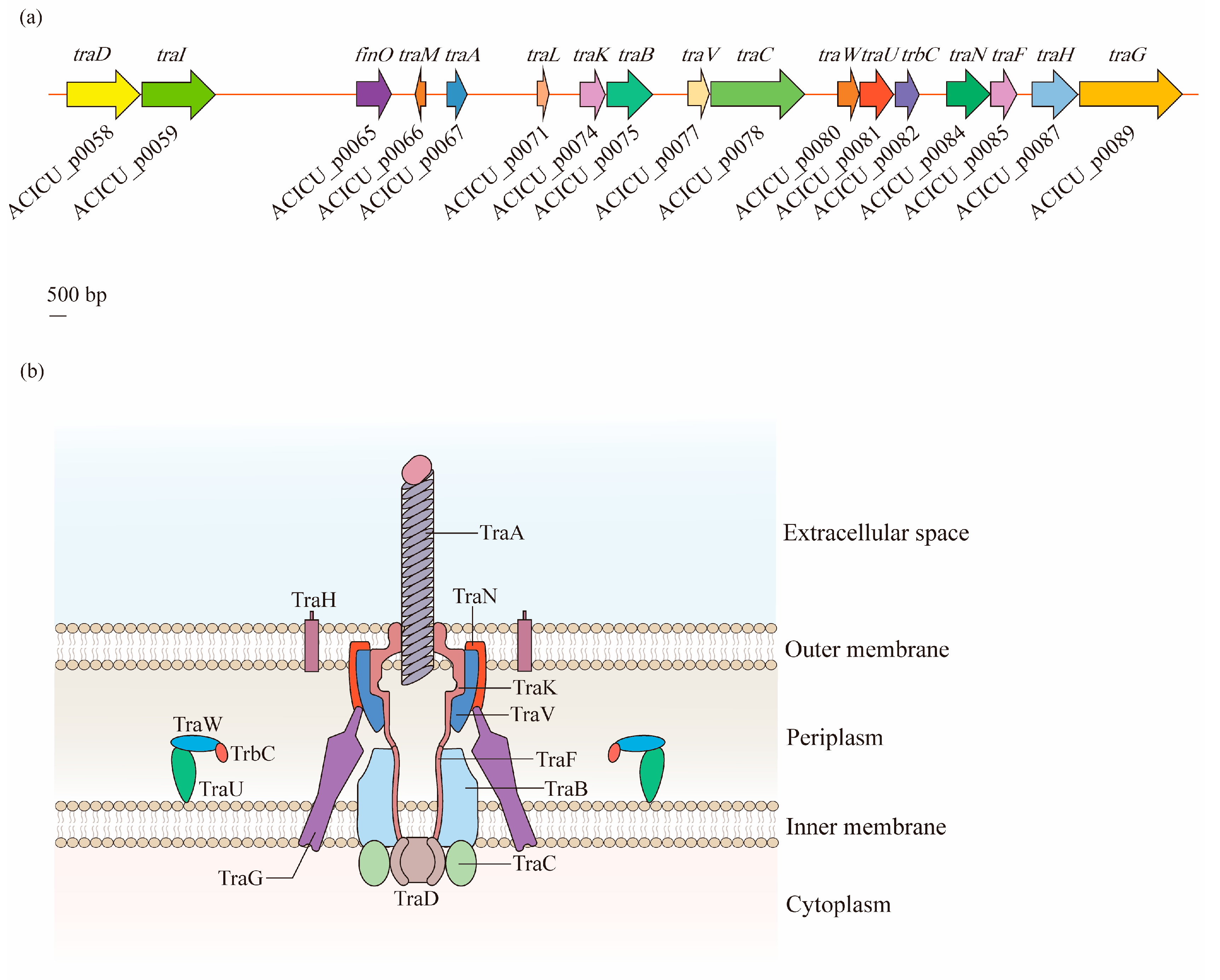
Figure 3. Structural organization of the type IV secretion system (T4SS) in A. baumannii: (a) Discovered in the A. baumannii ACICU plasmid pACICU2, the F-type T4SS contains a series of tra operon genes, and two other genes, trbC and finO. (b) The T4SS is a highly sophisticated nanomachine spanning the entire bacterial cell envelope in A. baumannii. The F-like T4SS apparatus is composed of a pilus assembly component (TraA), a core complex (TraK, TraV, TraN, and TraH) embedded in the outer membrane (OM), an inner membrane (IM) platform (TraF, TraB, TraG, TraU, TraW, and TrbC), and components of the cytoplasm (TraC and TraD).
According to the analysis of Liu et al. [43], the T4SS of A. baumannii is a symmetrical barrel-shaped structure that is divided into the following units: (1) the pilus assembly component localized in the extracellular space across the OM (TraA); (2) the core complex embedded in the OM (TraK, TraV, TraN, and TraH); (3) the constituents of an IM platform (TraF, TraB, TraG, TraU, TraW, and TrbC); and (4) the components of the cytoplasm (TraC and TraD). This structure is similar to that of the typical VirB/D4 T4SS, which exists on the Agrobacterium tumefaciens Ti plasmid, and has gene consistency with tra operons as traB/virB10, traC/virB4, and traD/virD4 [36][44] (Figure 3b).
5. Type V Secretion System (T5SS)
The T5SS, also known as the autotransporter, is a series of simple protein export pathways that are distributed in a large range of Gram-negative bacteria [45]. They are classified into monomeric autotransporters (MA), trimeric autotransporters (TA), and two-partner secretion systems (TPSS), with the composition of a single polypeptide for MA and TA, and separate polypeptide chains for TPSS [46][47]. Depending on the different structural features and domain organization, the T5SS is divided into five known subclasses, so-called types Va to Ve, and possibly another recently identified type, Vf [47]. However, only two types, Vb and Vc, have been identified in A. baumannii [27].
Gene and Structure
In contrast to other types of secretion systems that span the entire cell envelope with a syringe-shape structure, the T5SS only spans the OM. The T5SS consists of three major regions; namely, a signal sequence at the N-terminus, an extracellular secreted passenger, and a β-barrel domain (transporter) at the C-terminal that anchors the protein to the bacterial OM [47][48] (Figure 4a). Being produced in the cytoplasm, the protein is recognized at the N-terminal signal peptide, which targets the Sex complex to mediate the inner-membrane translocation of the protein to the periplasm [27]. Thereafter, the C-terminal transporter domain inserts into the OM and secretes the protein to the external environment through its OM pore. Finally, the passenger domain located between the signal peptide and the β-barrel domain displays the specific effector function extracellularly after proteolytic cleavage [33].
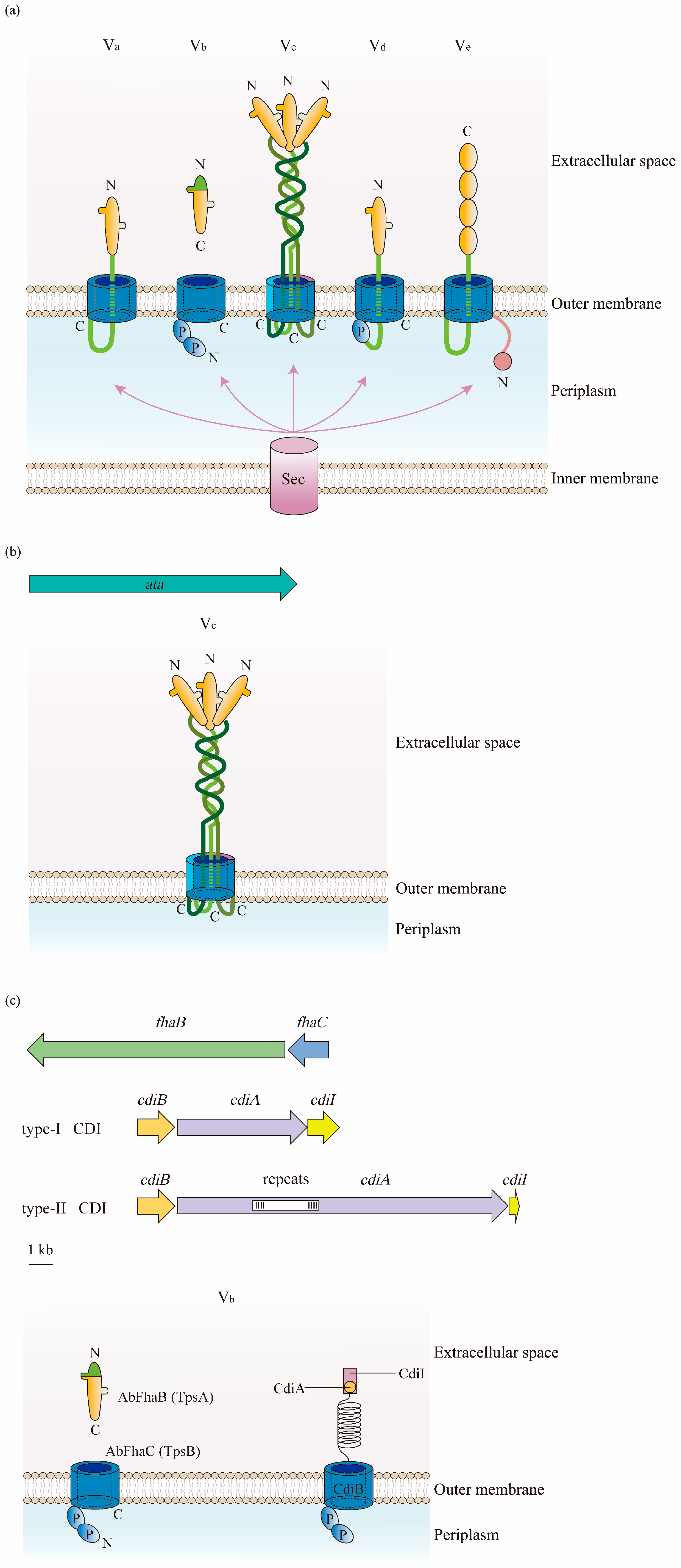
Figure 4. Structure of the type V secretion system (T5SS) in A. baumannii: (a) There are five types of T5SS in Gram-negative bacteria. They consist of three parts: a signal sequence at the N-terminus, a secreted passenger in the extracellular milieu, and a transporter at the C-terminal. β-Barrels are displayed in blue; linkers and the two-partner secretion (TPS) domains are in green; passenger regions are in orange; polypeptide transport-associated (POTRA) domains are labeled as P; and the N- and C-termini are indicated. The translocation of substrates for subclasses of T5SS from the cytoplasm to the periplasm relies on the Sec pathway. (b) Type Vc is the most frequently identified T5SS in A. baumannii. It is formed by a trimeric protein, Ata, which contains a signal peptide at the N-terminus, a surface-exposed passenger domain, and a C-terminal domain. (c) Two forms of type Vb are found in A. baumannii. The one belonging to the TPSS is constructed of AbFhaB and AbFhaC, which represent TpsA and TpsB in other Gram-negative bacteria, respectively. AbFhaB (TpsA) is the passenger domain that is secreted out of cells through the outer membrane (OM) by AbFhaC (TpsB), which is the translocator domain located in the OM. Another one is the contact-dependent inhibition (CDI) system composed of CdiA and CdiB. Similar to TpsA, the toxin CdiA is released from the periplasm to the cell surface by the OM transporter CdiB.
Type Vc is the most popular T5SS in the A. baumannii chromosome that belongs to the TA family. Therefore, the protein of type Vc in this bacterium is designated as the Acinetobacter trimeric autotransporter (Ata) [49]. Encoded by the ata gene, the autotransporter Ata contains a long signal peptide followed by an N-terminus, a surface-exposed passenger domain, and a C-terminal domain encoding four β-strands [49] (Figure 4b).
In contrast to classical autotransporters, type Vb belongs to TPSS, where the passenger and translocator (β-barrel) domains locate in two distinct polypeptide chains that are formed by TpsA and TpsB [46]. TpsA and TpsB are encoded in one operon, and the former connects at the polypeptide transport-associated (POTRA) domain of the latter for secretion through the OM to either be surface-displayed or transported extracellularly [50]. In this way, when releasing the passenger out of the cells after being transported by the β-barrel domain, there is no need for release by proteolytic cleavage [47]. In the A. baumannii strain AbH12O-A2, AbFhaB and AbFhaC were found to represent TpsA and TpsB, respectively, due to the highly conserved structure of these proteins [51] (Figure 4c).
Another type of Vb recently observed in A. baumannii is the CDI system composed of CdiA and CdiB. CdiA is a large multi-domain protein that forms a filament folded as a β-helix, similarly to TpsA, and has a C-terminal toxin domain. The CdiA protein in the periplasm is released to the cell surface by the OM transporter CdiB, and its β-helix presents the toxin domain to the neighboring bacteria, finally inhibiting their growth [52]. A cytoplasmic immunity protein, CdiI, is also expressed by the CDI operon to protect bacteria from fratricide and auto-inhibition by CdiA toxins [53][54][55] (Figure 4c).
6. Type VI Secretion System (T6SS)
The T6SS is a multiprotein transmembrane nanomachine discovered in numerous Gram-negative bacteria, including Vibrio cholerae, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Francisella tularensis, and Yersinia pseudotuberculosis [56]. It is syringe-shaped and is commonly used by bacteria to inject toxic effectors into competitors or host cells [57]. Several parts of this secretion system are structurally and functionally homologous to the T4 bacteriophage tail, suggesting a common evolutionary origin of this apparatus [58]. In recent years, an increasing number of studies have reported various aspects of the T6SS from A. baumannii, including its composition, structure, regulation, and function, confirming it as an important virulence factor.
Gene and Structure
The T6SS in A. baumannii is found in a cluster located in the genome that contains 18 genes, arranged as asaA-tssBC-hcp(tssD)-tssEFG-asaB-tssM-tagFN-asaC-tssHAKL-asaDE, while genes of vgrG, also known as tssI, which are scattered in various numbers throughout the genome [59][60]. In these genes, 12 encode the core T6SS proteins (Tss, Hcp, and VgrG), two encode the TagF and TagN that are associated with the T6SS in other bacteria, and five encode the Asa proteins that only appear in Acinetobacter spp. [59] (Figure 5a). Based on the Tss core proteins, the T6SS is composed of three main parts: a membrane complex, a cytoplasmic baseplate, and a contractile tail tube/sheath complex (Figure 5b).
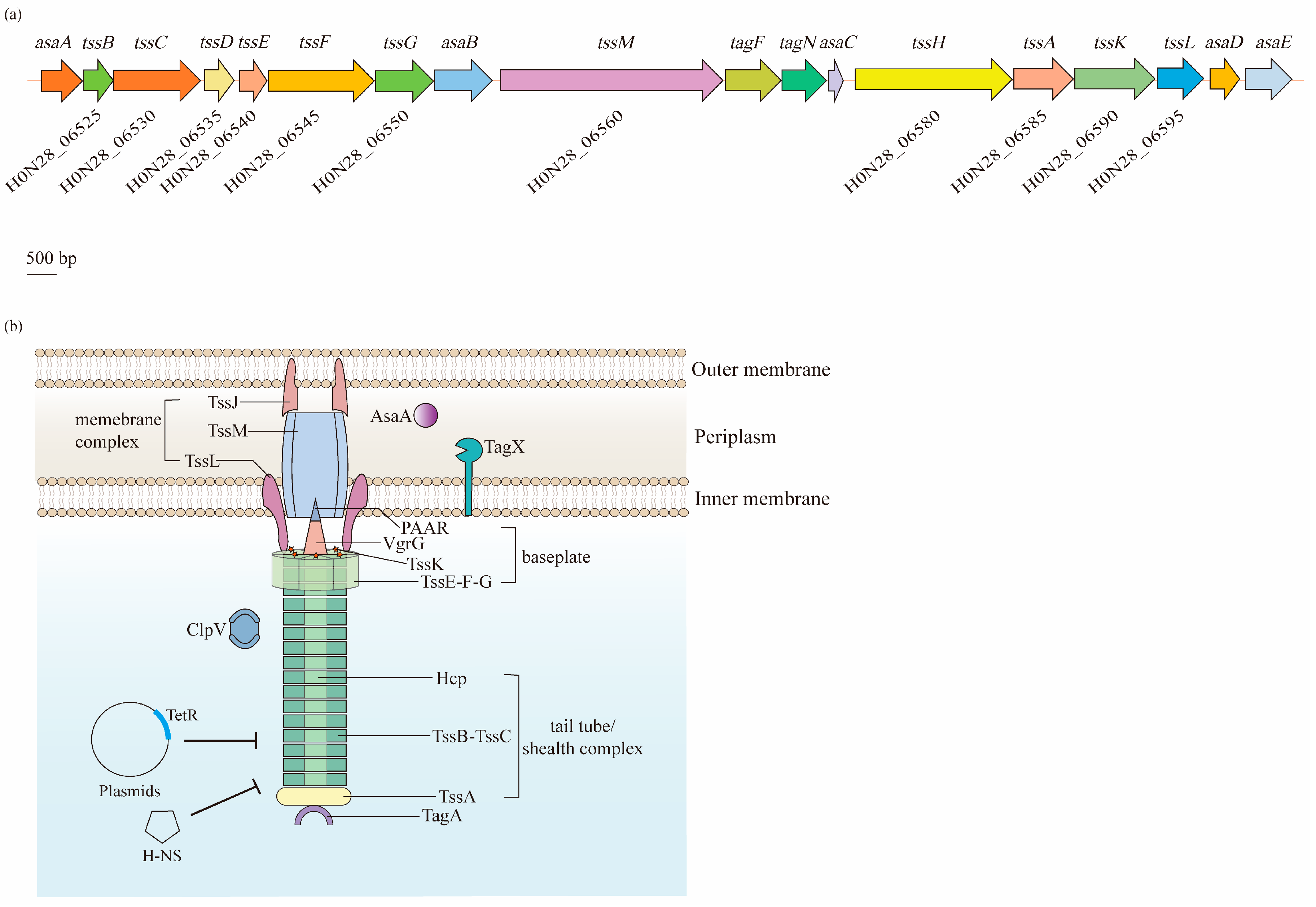
Figure 5. Biogenesis and regulation of the type VI secretion system (T6SS) in A. baumannii. The T6SS is a class of macromolecular secretion machines, which translocate proteins into a variety of recipient cells: (a) A single gene cluster carries 18 putative genes that are predicted to encode components of the T6SS. Among them, 12 core genes (tss) are coded on the chromosome of A. baumannii ATCC 17978. (b) The T6SS is composed of three main parts: a membrane complex (TssJ, TssL, and TssM), a cytoplasmic baseplate (TssK, TssF, TssG, TssE, VgrG, and PAAR), and a contractile tail tube/sheath complex (Hcp, TssB, TssC, and TssA). The expression of the T6SS is negatively regulated by the TetR-like proteins encoded on the large, conjugative plasmid pAB3 and proteins within the H-NS family.
Normally, in a wide range of bacteria, the membrane complex consists of the TssJ, TssL, and TssM proteins that span the cell envelope, with the complex anchored in the IM and the tip embedded in the OM, but not crossing it [61]. Notably, TssJ, an OM lipoprotein interacting with TssM, is absent in A. baumannii [60]. TssM and TssL have strong homology with the T4bSS proteins IcmF and IcmH (or DotU), respectively [62][63]. TssM is a core component of the T6SS that anchors to the IM through three transmembrane segments [63]. Similarly, the cytoplasmic protein TssL is also bound to the IM, but through a single transmembrane helix. Two residues of TssL in A. baumannii, Asp98 and Glu99, are strongly conserved among T6SS-encoding Gram-negative bacteria, and remarkably impact the dynamics, expression, and functionality of this protein [64]. TssM and TssL are involved in the recruitment and secretion of Hcp, and are important for the activity of the T6SS [65].
The baseplate complex is a central piece of the T6SS machinery that consists of six (TssK)6-(TssF)2-(TssG)1-(TssE)1 wedges around a central (VgrG)3-PAAR spike. It connects the tail to the membrane complex and initiates the polymerization of the tail tube/sheath complex [66]. TssG is the core component of a baseplate wedge, where its C-terminal domain acts as an adaptor to interact with both TssF and TssK. VgrG, which binds to the PAAR-repeat protein at its distal extremity, is essential for the assembly of the Hcp tube, thus significantly contributing to the structure of the T6SS in various bacteria, including A. baumannii [67][68][69].
The tail tube/sheath complex is a contractile structure formed by the Hcp tube, TssBC sheath, and TssA cap. Although VgrG locates in the center of the baseplate complex, it is identified as an extension of the Hcp tube, as the central density of the latter is uniform from the first ring docked on top of the (VgrG)3-PAAR spike [68]. Normally, the inner Hcp tube assembles onto the base of VgrG and extends into the cytoplasm. Simultaneously, the TssBC helical sheath polymerizes around the Hcp tube in an extended, high-energy “primed” conformation [70]. Additionally, its proximal ring has been suggested to interact with the TssK-TssF-TssG complex [67]. After contraction, the sheath is disassembled by the AAA+ ATPase ClpV for a new assembly cycle of an extended sheath [71]. Lastly, TssA is involved in the assembly of Hcp-TssBC, and caps the distal end of this structure [70][72].
In addition to the core components, additional auxiliaries are required for the A. baumannii T6SS to ensure the correct assembly and full activity. For example, TagF and TagN were identified to negatively regulate the activity of the T6SS, where the absence of these two proteins increased the secretion of Hcp [73]. Moreover, AsaA was demonstrated to localize in the periplasmic space and affect the assembly or stability of the T6SS by interacting with TssM [74]. Additionally, a novel peptidoglycan hydrolase, TagX, was proposed to be required for the transit of the T6SS machinery across the peptidoglycan layer, thus finally allowing the assembly of the T6SS [73].
References
- Giamarellou, H.; Antoniadou, A.; Kanellakopoulou, K. Acinetobacter baumannii: A universal threat to public health? Int. J. Antimicrob. Agents 2008, 32, 106–119.
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720.
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12.
- Pogue, J.M.; Kaye, K.S.; Cohen, D.A.; Marchaim, D. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin. Microbiol. Infect. 2015, 21, 302–312.
- Espinal, P.; Marti, S.; Vila, J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J. Hosp. Infect. 2012, 80, 56–60.
- Longo, F.; Vuotto, C.; Donelli, G. Biofilm formation in Acinetobacter baumannii. New Microbiol. 2014, 37, 119–127.
- Harris, A.D.; Johnson, J.K.; Pineles, L.; O’Hara, L.M.; Bonomo, R.A.; Thom, K.A. Patient-to-Patient Transmission of Acinetobacter baumannii Gastrointestinal Colonization in the Intensive Care Unit. Antimicrob. Agents Chemother. 2019, 63, e00392-19.
- Bayuga, S.; Zeana, C.; Sahni, J.; Della-Latta, P.; El-Sadr, W.; Larson, E. Prevalence and antimicrobial patterns of Acinetobacter baumannii on hands and nares of hospital personnel and patients: The iceberg phenomenon again. Heart Lung 2002, 31, 382–390.
- Nasr, P. Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter baumannii. J. Hosp. Infect. 2020, 104, 4–11.
- Nowak, P.; Paluchowska, P. Acinetobacter baumannii: Biology and drug resistance—Role of carbapenemases. Folia Histochem. Cytobiol. 2016, 54, 61–74.
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55.
- Gheorghe, I.; Barbu, I.C.; Surleac, M.; Sarbu, I.; Popa, L.I.; Paraschiv, S.; Feng, Y.; Lazar, V.; Chifiriuc, M.C.; Otelea, D.; et al. Subtypes, resistance and virulence platforms in extended-drug resistant Acinetobacter baumannii Romanian isolates. Sci. Rep. 2021, 11, 13288.
- Smiline Girija, A.S.; Ganesh, P.S. Virulence of Acinetobacter baumannii in proteins moonlighting. Arch. Microbiol. 2021, 204, 96.
- Wong, D.; Chao, J.D.; Av-Gay, Y. Mycobacterium tuberculosis-secreted phosphatases: From pathogenesis to targets for TB drug development. Trends Microbiol. 2013, 21, 100–109.
- Gerlach, R.G.; Hensel, M. Protein secretion systems and adhesins: The molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 2007, 297, 401–415.
- Sawa, T.; Shimizu, M.; Moriyama, K.; Wiener-Kronish, J.P. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: A review. Crit. Care 2014, 18, 668.
- Boudaher, E.; Shaffer, C.L. Inhibiting bacterial secretion systems in the fight against antibiotic resistance. Medchemcomm 2019, 10, 682–692.
- Ding, M.; Ye, Z.; Liu, L.; Wang, W.; Chen, Q.; Zhang, F.; Wang, Y.; Sjoling, A.; Martin-Rodriguez, A.J.; Hu, R.; et al. Subinhibitory antibiotic concentrations promote the horizontal transfer of plasmid-borne resistance genes from Klebsiellae pneumoniae to Escherichia coli. Front. Microbiol. 2022, 13, 1017092.
- Harding, C.M.; Pulido, M.R.; Di Venanzio, G.; Kinsella, R.L.; Webb, A.I.; Scott, N.E.; Pachon, J.; Feldman, M.F. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J. Biol. Chem. 2017, 292, 9075–9087.
- Sycz, G.; Di Venanzio, G.; Distel, J.S.; Sartorio, M.G.; Le, N.H.; Scott, N.E.; Beatty, W.L.; Feldman, M.F. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLoS Pathog. 2021, 17, e1009802.
- Ho, T.D.; Davis, B.M.; Ritchie, J.M.; Waldor, M.K. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect. Immun. 2008, 76, 1858–1865.
- Baldi, D.L.; Higginson, E.E.; Hocking, D.M.; Praszkier, J.; Cavaliere, R.; James, C.E.; Bennett-Wood, V.; Azzopardi, K.I.; Turnbull, L.; Lithgow, T.; et al. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect. Immun. 2012, 80, 2042–2052.
- McCoy-Simandle, K.; Stewart, C.R.; Dao, J.; DebRoy, S.; Rossier, O.; Bryce, P.J.; Cianciotto, N.P. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect. Immun. 2011, 79, 1984–1997.
- Sikora, A.E.; Zielke, R.A.; Lawrence, D.A.; Andrews, P.C.; Sandkvist, M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J. Biol. Chem. 2011, 286, 16555–16566.
- Jyot, J.; Balloy, V.; Jouvion, G.; Verma, A.; Touqui, L.; Huerre, M.; Chignard, M.; Ramphal, R. Type II secretion system of Pseudomonas aeruginosa: In vivo evidence of a significant role in death due to lung infection. J. Infect. Dis. 2011, 203, 1369–1377.
- Tomas, A.; Lery, L.; Regueiro, V.; Perez-Gutierrez, C.; Martinez, V.; Moranta, D.; Llobet, E.; Gonzalez-Nicolau, M.; Insua, J.L.; Tomas, J.M.; et al. Functional Genomic Screen Identifies Klebsiella pneumoniae Factors Implicated in Blocking Nuclear Factor kappaB (NF-kappaB) Signaling. J. Biol. Chem. 2015, 290, 16678–16697.
- Elhosseiny, N.M.; Attia, A.S. Acinetobacter: An emerging pathogen with a versatile secretome. Emerg. Microbes. Infect. 2018, 7, 33.
- Eijkelkamp, B.A.; Stroeher, U.H.; Hassan, K.A.; Paulsen, I.T.; Brown, M.H. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 2014, 15, 1020.
- Johnson, T.L.; Waack, U.; Smith, S.; Mobley, H.; Sandkvist, M. Acinetobacter baumannii Is Dependent on the Type II Secretion System and Its Substrate LipA for Lipid Utilization and In Vivo Fitness. J. Bacteriol. 2015, 198, 711–719.
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351.
- Thomassin, J.L.; Santos Moreno, J.; Guilvout, I.; Tran Van Nhieu, G.; Francetic, O. The trans-envelope architecture and function of the type 2 secretion system: New insights raising new questions. Mol. Microbiol. 2017, 105, 211–226.
- Harding, C.M.; Kinsella, R.L.; Palmer, L.D.; Skaar, E.P.; Feldman, M.F. Medically Relevant Acinetobacter Species Require a Type II Secretion System and Specific Membrane-Associated Chaperones for the Export of Multiple Substrates and Full Virulence. PLoS Pathog. 2016, 12, e1005391.
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359.
- Yan, Z.; Yin, M.; Xu, D.; Zhu, Y.; Li, X. Structural insights into the secretin translocation channel in the type II secretion system. Nat. Struct. Mol. Biol. 2017, 24, 177–183.
- Naskar, S.; Hohl, M.; Tassinari, M.; Low, H.H. The structure and mechanism of the bacterial type II secretion system. Mol. Microbiol. 2021, 115, 412–424.
- Costa, T.R.D.; Harb, L.; Khara, P.; Zeng, L.; Hu, B.; Christie, P.J. Type IV secretion systems: Advances in structure, function, and activation. Mol. Microbiol. 2021, 115, 436–452.
- Christie, P.J.; Atmakuri, K.; Krishnamoorthy, V.; Jakubowski, S.; Cascales, E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005, 59, 451–485.
- Juhas, M.; Crook, D.W.; Dimopoulou, I.D.; Lunter, G.; Harding, R.M.; Ferguson, D.J.; Hood, D.W. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 2007, 189, 761–771.
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614.
- Iacono, M.; Villa, L.; Fortini, D.; Bordoni, R.; Imperi, F.; Bonnal, R.J.; Sicheritz-Ponten, T.; De Bellis, G.; Visca, P.; Cassone, A.; et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 2008, 52, 2616–2625.
- Povilonis, J.; Seputiene, V.; Krasauskas, R.; Juskaite, R.; Miskinyte, M.; Suziedelis, K.; Suziedeliene, E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013, 68, 1000–1006.
- Towner, K.J.; Evans, B.; Villa, L.; Levi, K.; Hamouda, A.; Amyes, S.G.; Carattoli, A. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D beta-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 2154–2159.
- Liu, C.C.; Kuo, H.Y.; Tang, C.Y.; Chang, K.C.; Liou, M.L. Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics 2014, 104, 215–223.
- Juhas, M.; Crook, D.W.; Hood, D.W. Type IV secretion systems: Tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 2008, 10, 2377–2386.
- Bernstein, H.D. Type V Secretion in Gram-Negative Bacteria. EcoSal Plus 2019, 8.
- Leo, J.C.; Grin, I.; Linke, D. Type V secretion: Mechanism(s) of autotransport through the bacterial outer membrane. Philos. Trans. R. Soc. Lond B Biol. Sci. 2012, 367, 1088–1101.
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V Secretion Systems: An Overview of Passenger Domain Functions. Front. Microbiol. 2019, 10, 1163.
- Jose, J.; Jahnig, F.; Meyer, T.F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol. Microbiol. 1995, 18, 378–380.
- Bentancor, L.V.; Camacho-Peiro, A.; Bozkurt-Guzel, C.; Pier, G.B.; Maira-Litran, T. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 2012, 194, 3950–3960.
- Thanassi, D.G.; Stathopoulos, C.; Karkal, A.; Li, H. Protein secretion in the absence of ATP: The autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria (review). Mol. Membr. Biol. 2005, 22, 63–72.
- Perez, A.; Merino, M.; Rumbo-Feal, S.; Alvarez-Fraga, L.; Vallejo, J.A.; Beceiro, A.; Ohneck, E.J.; Mateos, J.; Fernandez-Puente, P.; Actis, L.A.; et al. The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 2017, 8, 959–974.
- Guerin, J.; Botos, I.; Zhang, Z.; Lundquist, K.; Gumbart, J.C.; Buchanan, S.K. Structural insight into toxin secretion by contact-dependent growth inhibition transporters. Elife 2020, 9, e58100.
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D.A. Contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309, 1245–1248.
- Ruhe, Z.C.; Subramanian, P.; Song, K.; Nguyen, J.Y.; Stevens, T.A.; Low, D.A.; Jensen, G.J.; Hayes, C.S. Programmed Secretion Arrest and Receptor-Triggered Toxin Export during Antibacterial Contact-Dependent Growth Inhibition. Cell 2018, 175, 921–933.
- Ruhe, Z.C.; Nguyen, J.Y.; Xiong, J.; Koskiniemi, S.; Beck, C.M.; Perkins, B.R.; Low, D.A.; Hayes, C.S. CdiA Effectors Use Modular Receptor-Binding Domains To Recognize Target Bacteria. mBio 2017, 8, e00290-17.
- Monjaras Feria, J.; Valvano, M.A. An Overview of Anti-Eukaryotic T6SS Effectors. Front. Cell. Infect. Microbiol. 2020, 10, 584751.
- Hofer, U. T6SS: Shoot and scrub. Nat. Rev. Microbiol. 2020, 18, 412–413.
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159.
- Carruthers, M.D.; Nicholson, P.A.; Tracy, E.N.; Munson, R.S., Jr. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS ONE 2013, 8, e59388.
- Weber, B.S.; Miyata, S.T.; Iwashkiw, J.A.; Mortensen, B.L.; Skaar, E.P.; Pukatzki, S.; Feldman, M.F. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS ONE 2013, 8, e55142.
- Rapisarda, C.; Cherrak, Y.; Kooger, R.; Schmidt, V.; Pellarin, R.; Logger, L.; Cascales, E.; Pilhofer, M.; Durand, E.; Fronzes, R. In Situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J. 2019, 38, e100886.
- Nguyen, V.S.; Douzi, B.; Durand, E.; Roussel, A.; Cascales, E.; Cambillau, C. Towards a complete structural deciphering of Type VI secretion system. Curr. Opin. Struct. Biol. 2018, 49, 77–84.
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 2012, 66, 453–472.
- Ruiz, F.M.; Lopez, J.; Ferrara, C.G.; Santillana, E.; Espinosa, Y.R.; Feldman, M.F.; Romero, A. Structural Characterization of TssL from Acinetobacter baumannii: A Key Component of the Type VI Secretion System. J. Bacteriol. 2020, 202, e00210-20.
- Ma, L.S.; Narberhaus, F.; Lai, E.M. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 2012, 287, 15610–15621.
- Cherrak, Y.; Rapisarda, C.; Pellarin, R.; Bouvier, G.; Bardiaux, B.; Allain, F.; Malosse, C.; Rey, M.; Chamot-Rooke, J.; Cascales, E.; et al. Biogenesis and structure of a type VI secretion baseplate. Nat. Microbiol. 2018, 3, 1404–1416.
- Park, Y.J.; Lacourse, K.D.; Cambillau, C.; DiMaio, F.; Mougous, J.D.; Veesler, D. Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy. Nat. Commun. 2018, 9, 5385.
- Nazarov, S.; Schneider, J.P.; Brackmann, M.; Goldie, K.N.; Stahlberg, H.; Basler, M. Cryo-EM reconstruction of Type VI secretion system baseplate and sheath distal end. EMBO J. 2018, 37, e97103.
- Lopez, J.; Ly, P.M.; Feldman, M.F. The Tip of the VgrG Spike Is Essential to Functional Type VI Secretion System Assembly in Acinetobacter baumannii. mBio 2020, 11, e02761-19.
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515.
- Forster, A.; Planamente, S.; Manoli, E.; Lossi, N.S.; Freemont, P.S.; Filloux, A. Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J. Biol. Chem. 2014, 289, 33032–33043.
- Dix, S.R.; Owen, H.J.; Sun, R.; Ahmad, A.; Shastri, S.; Spiewak, H.L.; Mosby, D.J.; Harris, M.J.; Batters, S.L.; Brooker, T.A.; et al. Structural insights into the function of type VI secretion system TssA subunits. Nat. Commun. 2018, 9, 4765.
- Weber, B.S.; Hennon, S.W.; Wright, M.S.; Scott, N.E.; de Berardinis, V.; Foster, L.J.; Ayala, J.A.; Adams, M.D.; Feldman, M.F. Genetic Dissection of the Type VI Secretion System in Acinetobacter and Identification of a Novel Peptidoglycan Hydrolase, TagX, Required for Its Biogenesis. mBio 2016, 7, e01253-16.
- Li, L.; Wang, Y.N.; Jia, H.B.; Wang, P.; Dong, J.F.; Deng, J.; Lu, F.M.; Zou, Q.H. The type VI secretion system protein AsaA in Acinetobacter baumannii is a periplasmic protein physically interacting with TssM and required for T6SS assembly. Sci. Rep. 2019, 9, 9438.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.5K
Revisions:
2 times
(View History)
Update Date:
02 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




