
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Karolina Wleklik | -- | 1667 | 2023-01-26 11:42:33 | | | |
| 2 | Sławomir Borek | + 222 word(s) | 1947 | 2023-01-30 15:34:30 | | | | |
| 3 | Catherine Yang | -58 word(s) | 1889 | 2023-01-31 02:01:50 | | | | |
| 4 | Catherine Yang | Meta information modification | 1889 | 2023-01-31 02:02:07 | | |
Video Upload Options
Autophagy is considered as a two-faced process: it can ensure cell survival as well as promote cell death. Autophagic cell death (ACD) is the second form of animal PCD. It is associated with increased numbers of autophagosomes, autolysosomes, and small lytic vacuoles. Autophagic death is a controversial idea that has been discussed and debated many times. Although our knowledge of this subject in plants is limited, a few examples of cell death with autophagy have been described.
1. Autophagy - a housekeeping process in plant cell
Autophagy is the evolutionarily well-conserved process of cell self-eating occurring in yeasts, animals, and plants. Through the autophagy pathway, cellular components such as protein complexes and organelles are degraded. Moreover, bacteria and viruses can also be degraded in the infected cells through this process [1]. Autophagy takes place in all life stages of the plant, including development, senescence, and cell death [2][3]. Under normal development and growth conditions, the insensitivity of autophagy is relatively low—basal. Then, it works as a quality control mechanism to degrade and recycle unwanted or damaged cellular components [4]. However, it remarkably increases during biotic and abiotic stresses such as nutrient deficiency, drought, salinity, heat, oxidation, and pathogen attack [2]. In yeast, autophagy is regulated by over forty AuTophaGy-related (Atg) genes, which have also been found in animals and plants [5]. These genes encode Atg proteins, which play many roles during autophagy processes. For example, the Atg1/Atg13 kinase complex is essential for autophagy initiation by the target of rapamycin (TOR) signaling pathway [6]. TOR, a serine/threonine kinase, negatively regulates autophagy in response to many environmental stimuli [7], whereas the sucrose nonfermenting-1-related protein kinase 1 (SnRK1) is the central kinase complex, which positively regulates autophagy by activation of Atg1 kinase [8]. Autophagy occurs in both selective and non-selective ways. The selective form of autophagy takes place when only particular cell components are degraded, for example, mitochondria (mitophagy) or peroxisomes (pexophagy) [9][10][11]. Due to the differences in the delivery of the cargo intended for autophagic degradation, the following types of autophagy in plants are distinguished: macroautophagy, microautophagy, and mega-autophagy [12][13][14][15]. Macroautophagy (Figure 1a) is the best-known type of autophagy. It starts with the formation in the cytoplasm of a cup-shaped structure named a phagophore. The phagophore elongates until it is surrounded by cell components intended for degradation. A vesicle with a bilayer double-membrane, containing cargo intended for degradation, is called an autophagosome. These stages of macroautophagy are similar in yeasts, plants, and animals. The next stage, which is directing the autophagosome to the lytic cell compartments, is similar in yeasts and plants but distinguished from animals. Namely, in yeasts and plants, the autophagosome is directed to the vacuole, where it fuses with the tonoplast by its outer membrane. The unaffected internal membrane of the autophagosome with the cargo inside creates an autophagic body inside the vacuole. In animals, the autophagosome is directed to the lysosome, where they fuse, creating an autolysosome. Finally, in both the vacuole and the autolysosome, cargo is degraded by lytic enzymes [11][12][16]. Microautophagy (Figure 1b) is an autophagy pathway in which autophagosomes are not formed. Cell elements intended for degradation enter the lysosome or vacuole by membrane invagination or protrusion of these organelles [11][12]. Mega-autophagy (Figure 1c) has only been observed in plants, and it is perceived as massive cytoplasm destruction that occurs during dPCD and abiotic stress-induced ePCD. Nonetheless, none of the Atg genes are involved in mega-autophagy, and cellular components are not directed to the vacuole for degradation [17][18]
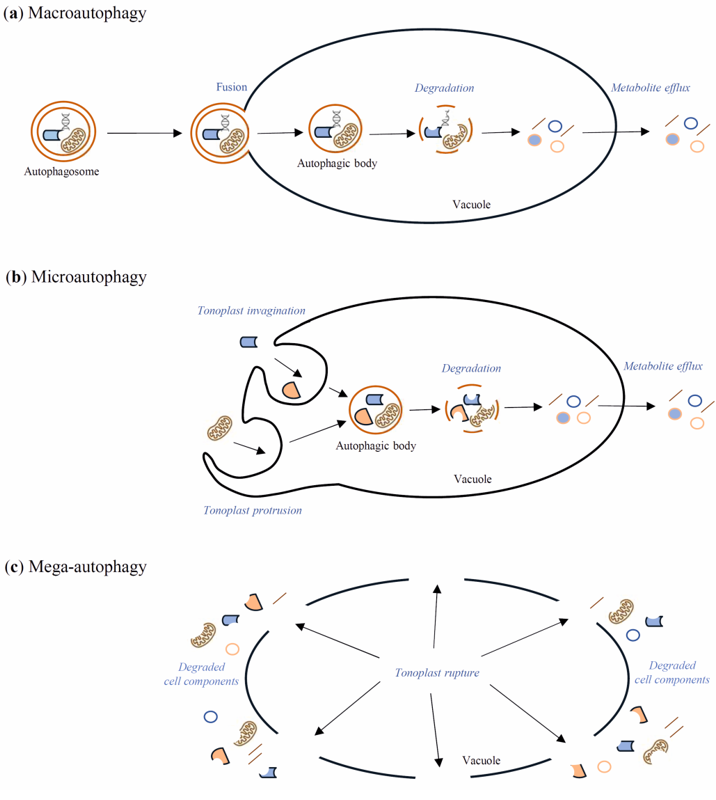
Figure 1. Schematic diagram of macroautophagy (a), microautophagy (b), and mega-autophagy (c) in plants. During macroautophagy, cargo intended for degradation is transported to the vacuole inside an autophagosome. The outer membrane of the autophagosome fuses with the tonoplast, while the internal autophagosome membrane and the cargo create an autophagic body inside the vacuole. The autophagic body is rapidly degraded by vacuolar hydrolases, which allow for the recycling of metabolites. During microautophagy, the autophagosome is not formed, but cell components intended for degradation enter the vacuole through the tonoplast invagination or tonoplast protrusion. Inside the vacuole, there arise the autophagic bodies, which, as in macroautophagy, are degraded by vacuolar hydrolases. Mega-autophagy differs significantly from macro- and microautophagy, as cell elements are not transported to the vacuole for degradation. Instead, the vacuole membrane is destroyed, and subsequently cell death occurs.
2. "Autophagic death" or "Death with autophagy"?
Autophagic cell death (ACD) is the second form of animal PCD. It is associated with increased numbers of autophagosomes, autolysosomes, and small lytic vacuoles [19]. Autophagic death is a controversial idea that has been discussed and debated many times. Autophagy is considered as a two-faced process: it can ensure cell survival as well as promote cell death [20]. However, it is difficult to distinguish when the occurrence of autophagic-related structures and recruitment of Atg genes function with the aim of cell survival and, conversely, when the aim is cell death. To solve this problem, it has been proposed to define “autophagic death” as when inhibition of autophagy contributes to long-term cell survival. In contrast, “cell death with autophagy” should be defined when inhibition of autophagy does not determine the subsequent death of the cell, but may change its morphology and delay the process (Figure 2) [21]. Therefore, crosstalk between these two processes remains important to study. Many genes are involved in both autophagy and cell death in animal models [20]. Although our knowledge of this subject in plants is limited, a few examples of cell death with autophagy have been described, and VPEs, as proteases associated with cell death execution in plants, may be an important factor connected with the pro-death or pro-survival role of plant autophagy [22][23].
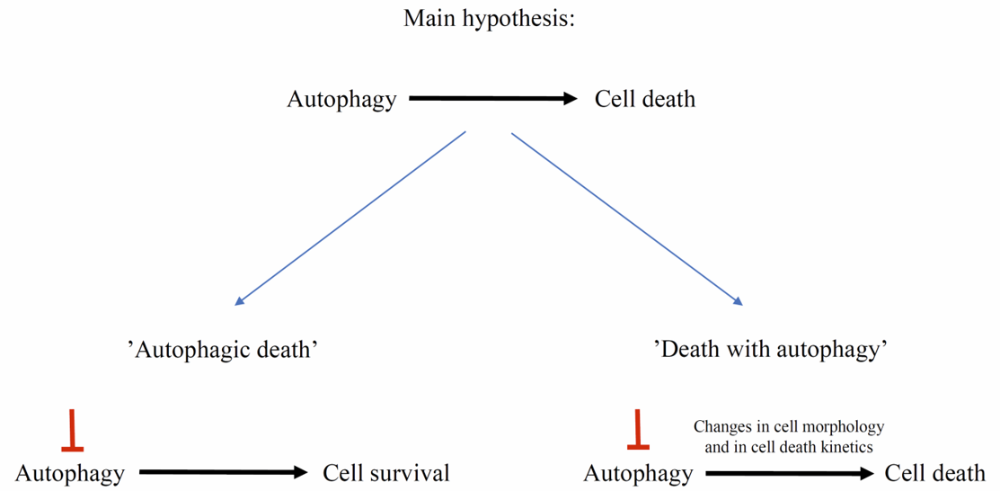
Figure 2. To determine whether autophagy acts in a cell pro-death or pro-survival manner, an experimental approach based on inhibition of autophagy is needed. “Autophagic death” occurs when inhibition of autophagy contributes to cell survival, whereas “death with autophagy” occurs when inhibition of autophagy, for example, delays cell death, but finally it will occur.
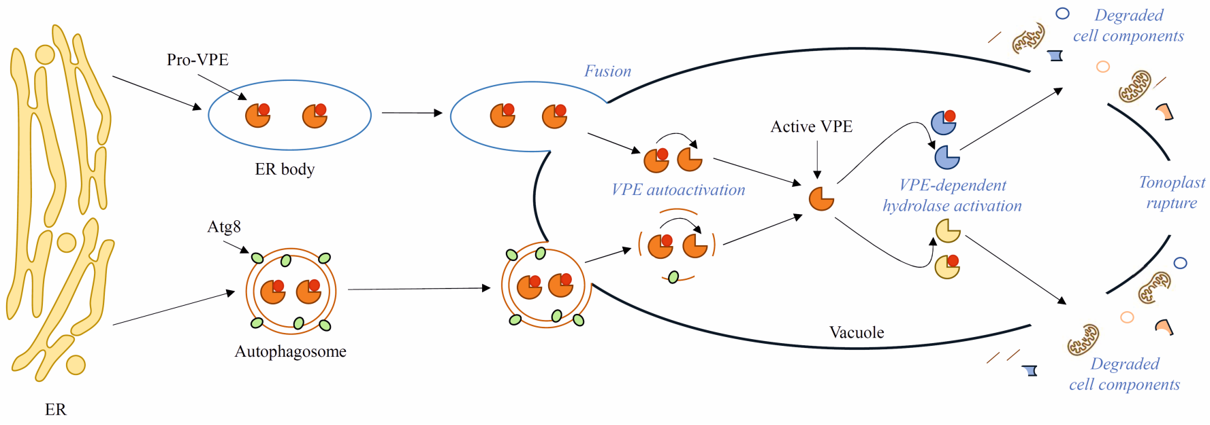
3. VPEs as a crosstalk point between autophagy and PCD
4. Conclusions and Future Perspectives
Autophagy was first observed in the 1950s [35]. Decades of research have revealed the key importance of autophagy in plant development and responses to internal and external stimuli. Nonetheless, many aspects, for example, the late stages of autophagy, i.e., degradation of autophagic bodies and metabolite efflux from the vacuole to the cytoplasm, have been overlooked in the research, and the knowledge about these stages of autophagy in plants is vestigial. The process of degradation of autophagic bodies in yeast occurs with the participation of several known enzymes, whereas in plants, only VPEs are taken into consideration as potentially involved in this process. However, it is not clear how VPEs would distinguish their pro-death activity during PCD from pro-survival activity during autophagy and limit their role only to the initiation of autophagic body degradation. Moreover, the links between autophagy and PCD are still poorly understood. The evidence that VPEs may be delivered to the vacuole by the autophagy pathway seems to be a good reference point for future investigations. The Atg8 gene family, which encodes ubiquitin-like proteins required for autophagosome formation, should also be examined as a potential point of crosstalk between autophagy and cell death.
References
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al.et al. Autophagy Functions as an Antiviral Mechanism against Geminiviruses in Plants. eLife 2017, 6, 23897.
- Yang, Y.; Xiang, Y.; Niu, Y. An Overview of the Molecular Mechanisms and Functions of Autophagic Pathways in Plants. Plant. Signal. Behav. 2021, 16, 1977527.
- Hashimi, S.M.; Wu, N.-N.; Ran, J.; Liu, J.-Z. Silencing Autophagy-Related Gene 2 (ATG2) Results in Accelerated Senescence and Enhanced Immunity in Soybean. Int. J. Mol. Sci. 2021, 22, 11749.
- Chen, H.; Dong, J.; Wang, T. Autophagy in Plant Abiotic Stress Management. Int. J. Mol. Sci. 2021, 22, 4075.
- Rehman, N.U.; Zeng, P.; Mo, Z.; Guo, S.; Liu, Y.; Huang, Y.; Xie, Q. Conserved and Diversified Mechanism of Autophagy between Plants and Animals upon Various Stresses. Antioxidants 2021, 10, 1736.
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 Protein Kinase Complex Is Both a Regulator and a Target of Autophagic Recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779.
- Fu, L.; Wang, P.; Xiong, Y. Target of Rapamycin Signaling in Plant Stress Responses. Plant Physiol. 2020, 182, 1613–1623.
- Cao, J.-J.; Liu, C.-X.; Shao, S.-J.; Zhou, J. Molecular Mechanisms of Autophagy Regulation in Plants and Their Applications in Agriculture. Front. Plant Sci. 2021, 11, 618944.
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biol. 2018, 20, 233–242.
- Stephani, M.; Dagdas, Y. Plant Selective Autophagy—Still an Uncharted Territory With a Lot of Hidden Gems. J. Mol. Biol. 2020, 432, 63–79.
- Borek, S.; Stefaniak, S.; Śliwiński, J.; Garnczarska, M.; Pietrowska-Borek, M. Autophagic Machinery of Plant Peroxisomes. Int. J. Mol. Sci. 2019, 20, 4754.
- Stefaniak, S.; Wojtyla, Ł.; Pietrowska-Borek, M.; Borek, S. Molecular Sciences Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants. Int. J. Mol. Sci. 2020, 21, 2205.
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164.
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466.
- Iglesias-Fernández, R.; Vicente-Carbajosa, J. A View into Seed Autophagy: From Development to Environmental Responses. Plants 2022, 11, 3247.
- Gomez, R.E.; Lupette, J.; Chambaud, C.; Castets, J.; Ducloy, A.; Cacas, J.-L.; Masclaux-Daubresse, C.; Bernard, A. How Lipids Contribute to Autophagosome Biogenesis, a Critical Process in Plant Responses to Stresses. Cells 2021, 10, 1272.
- van Doorn, W.G.; Papini, A. Ultrastructure of Autophagy in Plant Cells. Autophagy 2013, 9, 1922–1936.
- Wang, P.; Wang, T.; Han, J.; Li, M.; Zhao, Y.; Su, T.; Ma, C. Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways. Front. Plant Sci. 2021, 12, 754982.
- van Doorn, W.G. Classes of Programmed Cell Death in Plants, Compared to Those in Animals. J. Exp. Bot. 2011, 62, 4749–4761.
- Yu, G.; Klionsky, D. Life and Death Decisions-The Many Faces of Autophagy in Cell Survival and Cell Death. Biomolecules 2022, 12, 866.
- Kroemer, G.; Levine, B. Autophagic Cell Death: The Story of a Misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010.
- Li, J.-W.; Zhang, S.-B.; Xi, H.-P.; Bradshaw, C.J.A.; Zhang, J.-L. Processes Controlling Programmed Cell Death of Root Velamen Radicum in an Epiphytic Orchid. Ann. Bot. 2020, 126, 261–275.
- Teper-Bamnolker, P.; Danieli, R.; Peled-Zehavi, H.; Belausov, E.; Abu-Abied, M.; Avin-Wittenberg, T.; Sadot, E.; Eshel, D. Vacuolar Processing Enzyme Translocates to the Vacuole through the Autophagy Pathway to Induce Programmed Cell Death. Autophagy 2021, 17, 3109–3123.
- Kwon, S.I.; Cho, H.J.; Jung, J.H.; Yoshimoto, K.; Shirasu, K.; Park, O.K. The Rab GTPase RabG3b Functions in Autophagy and Contributes to Tracheary Element Differentiation in Arabidopsis. Plant J. 2010, 64, 151–164.
- Wojciechowska, N.; Michalak, K.M.; Bagniewska-Zadworna, A. Autophagy—An Underestimated Coordinator of Construction and Destruction during Plant Root Ontogeny. Planta 2021, 254, 15.
- Sobieszczuk-Nowicka, E.; Wrzesiński, T.; Bagniewska-Zadworna, A.; Kubala, S.; Rucińska-Sobkowiak, R.; Polcyn, W.; Misztal, L.; Mattoo, A.K. Physio-Genetic Dissection of Dark-Induced Leaf Senescence and Timing Its Reversal in Barley. Plant Physiol. 2018, 178, 654–671.
- Feng, Q.; De Rycke, R.; Dagdas, Y.; Nowack, M.K. Autophagy Promotes Programmed Cell Death and Corpse Clearance in Specific Cell Types of the Arabidopsis Root Cap. Curr. Biol. 2022, 32, 2110–2119.
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 Is Required to Limit the Pathogen-Associated Cell Death Response. Autophagy 2008, 4, 20–27.
- Hofius, D.; Schultz-Larsen, T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.T.; Mattsson, O.; Jørgensen, L.B.; Jones, J.D.G.; Mundy, J.; Petersen, M. Autophagic Components Contribute to Hypersensitive Cell Death in Arabidopsis. Cell 2009, 137, 773–783.
- Sertsuvalkul, N.; DeMell, A.; Dinesh-Kumar, S.P. The Complex Roles of Autophagy in Plant Immunity. FEBS Lett. 2022, 596, 2163–2171.
- Hayashi, Y.; Yamada, K.; Shimada, T.; Matsushima, R.; Nishizawa, N.; Nishimura, M.; Hara-Nishimura, I. A Proteinase-Storing Body That Prepares for Cell Death or Stresses in the Epidermal Cells of Arabidopsis. Plant Cell Physiol. 2001, 42, 894–899.
- Michaeli, S.; Avin-Wittenberg, T.; Galili, G. Involvement of Autophagy in the Direct ER to Vacuole Protein Trafficking Route in Plants. Front. Plant Sci. 2014, 5, 134.
- Li, Y.-B.; Yan, M.; Cui, D.-Z.; Huang, C.; Sui, X.-X.; Guo, F.Z.; Fan, Q.-Q.; Chu, X.-S. Programmed Degradation of Pericarp Cells in Wheat Grains Depends on Autophagy. Front. Genet. 2021, 12, 784545.
- Dauphinee, A.N.; Denbigh, G.L.; Rollini, A.; Fraser, M.; Lacroix, C.R.; Gunawardena, A.H.L.A.N. The Function of Autophagy in Lace Plant Programmed Cell Death. Front. Plant Sci. 2019, 10, 1198.
- Clark, S.L. Cellular Differentiation in the Kidneys of Newborn Mice Studies with the Electron Microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362.




