Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jack Kelly | -- | 2515 | 2023-01-17 04:46:23 | | | |
| 2 | Jack Kelly | Meta information modification | 2515 | 2023-01-17 04:53:03 | | | | |
| 3 | Rita Xu | Meta information modification | 2515 | 2023-01-17 06:09:50 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kelly, J.H.; Tucker, M.R.; Brewer, P.B. Strigolactone-Mediated Bud Outgrowth. Encyclopedia. Available online: https://encyclopedia.pub/entry/40261 (accessed on 08 February 2026).
Kelly JH, Tucker MR, Brewer PB. Strigolactone-Mediated Bud Outgrowth. Encyclopedia. Available at: https://encyclopedia.pub/entry/40261. Accessed February 08, 2026.
Kelly, Jack H., Matthew R. Tucker, Philip B. Brewer. "Strigolactone-Mediated Bud Outgrowth" Encyclopedia, https://encyclopedia.pub/entry/40261 (accessed February 08, 2026).
Kelly, J.H., Tucker, M.R., & Brewer, P.B. (2023, January 17). Strigolactone-Mediated Bud Outgrowth. In Encyclopedia. https://encyclopedia.pub/entry/40261
Kelly, Jack H., et al. "Strigolactone-Mediated Bud Outgrowth." Encyclopedia. Web. 17 January, 2023.
Copy Citation
Strigolactones (SLs), being a novel class of phytohormone, are known to play a key role in branching decisions, where they act as a negative regulator of bud outgrowth. They can achieve this by modulating polar auxin transport to interrupt auxin canalisation, and independently of auxin by acting directly within buds by promoting the key branching inhibitor TEOSINTE BRANCHED1.
shoot branching
bud outgrowth
strigolactone
shoot architecture
1. Introduction
The developmental blueprint of seed plants includes the phenomenal ability to adapt their architecture by growing new organs. Regulating branch number is an important component of plant development. Branches can be replicated almost ad infinitum by bud production and outgrowth. Axillary buds are formed in the axils of leaves. Some buds never cease development and grow into a branch. Others may cease development and enter a metabolically active but non-growing state in certain conditions. These buds may resume development later if required, while others may cease development altogether. The ability to perceive the environment and make these branching decisions is made possible by phytohormones that act within a highly complex signalling network, which allows for chemical signals to be perceived and a growth response to be triggered [1]. Strigolactones (SLs) are one of multiple important signals that play a key role in branching regulation, where they act as a decision for a bud to enter a non-growing state. By acting as a negative regulator of branching, they can modulate plant architecture to optimise growth. Findings have shown that SLs can inhibit bud outgrowth by promoting transcriptional inhibitors, and by modulating auxin transport to influence apical dominance [2].
2. Strigolactone Classification and Synthesis
SLs are a novel class of phytohormones comprised of several structurally diverse molecules that have been identified to regulate numerous aspects of plant function and development. This includes plant stature, inflorescence architecture, shade avoidance, root architecture, senescence and abiotic and biotic stress tolerance. SLs are also exuded from roots to influence soil microbe symbiosis and parasitic weed germination. Strigol was the first SL isolated from root exudates in cotton, where it was identified as a germination stimulant for the parasitic Striga lutea [3]. Ongoing technological advancements have since led to the characterisation of over 30 SLs, with some being widely distributed across plant genera and others being specific to the species from which they were isolated [4]. SLs can be defined as canonical or non-canonical subject to key differences in their chemical structure, with canonical SLs consisting of a tricyclic lactone core (ABC ring) that is connected to a butanolide moiety (D ring) via an enol ether bond [5]. Orobanchol is another SL that was later identified as a germination stimulant for the parasite Orobanche minor, with subsequent stereochemical analysis identifying an α-orientated C-ring unlike the β-orientated C-ring seen in strigol-type SLs [6][7]. Since this discovery, newly identified canonical SLs have been classified as strigol-like or obobancol-like subject to the orientation of the C-ring. In contrast, non-canonical SLs lack elements of the conventional A-, B- and C-ring structure, but retain the D-ring (hydroxymethylbutenolide), which is essential for SL activity [8].
Early studies identified that SL stimulants are carotenoid derived when analysing carotenoid deficient hosts. Subsequent genetic screening of shoot branching mutants identified β-carotene isomerase and carotenoid cleavage dioxygenases CCD7 and CCD8 as key enzymes that catalyse the synthesis of SL molecules (Figure 1) [9][10][11]. The discovery of these enzymes allowed for the initial stages of SL biosynthesis to be outlined. The all-trans-β-carotene precursor is converted to 9-cis-β-carotene by the β-carotene isomerase, which is then cleaved by CCD7 into 9-cis-β-apo-10′-carotenal. CCD8 then converts this to carlactone (CL). CL has the A- and D-ring structure and was later identified to be an endogenous precursor for both canonical and non-canonical SLs [12]. CYP711A1, a cytochrome P450 monooxygenase (CYP450), was first identified in Arabidopsis (Arabidopsis thaliana) to act downstream of CCD7 and CCD8 in the SL biosynthesis pathway [13]. It was subsequently found that CYP711A1 catalyses three oxidation reactions that convert CL to carlactonoic acid (CLA) and that this conversion is conserved in vascular plants [14][15]. CLA is important as it acts as the precursor for all known SLs, including 5-deoxystrigol (5DS) and 4-deoxyorobanchol (4DO), which are the precursors for strigol and orobanchol-like SLs [16]. Other conversions downstream of CYP711A1, by a range of CYP450s and other enzymes, facilitate the diversity and species specificity of SLs [4]. One of these conversions involves CLA methyltransferase (CLAMT), which converts CLA to methyl carlactonoate (MeCLA) [17]. Another enzyme, known as LATERAL BRANCHING OXIDOREDUCTASE (LBO), then catalyses a further conversion of MeCLA to 1′OH-MeCLA, while also demethylating MeCLA to produce CLA [17][18]. This highlights that LBO is likely a key enzyme for SL diversity, although many aspects of its function along with other conversions in the pathway remain poorly understood. While it is known that different SL types have varied bioactivity, the underlying mechanisms have not been discovered [4]. Elucidating these unknown mechanisms in the SL biosynthesis pathway will be important for developing new variants for investigating the influence of this hormone on plant development and response, and the impact of different SLs in the rhizosphere. Additionally, it has also been observed that carboxylesterase enzymes (CXEs) are involved in SL catabolism and sequestration (Figure 1) [19][20]. Context-specific enzyme gene expression and transport of SLs may also be important for function. Rice CYP450s show distinctive expression responses, and PLEIOTROPIC DRUG RESISTANCE 1 (PDR1) has been identified as a polar transporter in petunia that facilitates short-distance SL transport internally in the plant and out into the rhizosphere [21][22]. Rice plants mutated in a specific CYP450 (Os900) failed to show root exudation, but retained normal branching, indicating a possible biosynthesis pathway specific for root exudation, although further analysis of individual biosynthesis genes is required to unravel these effects [23].
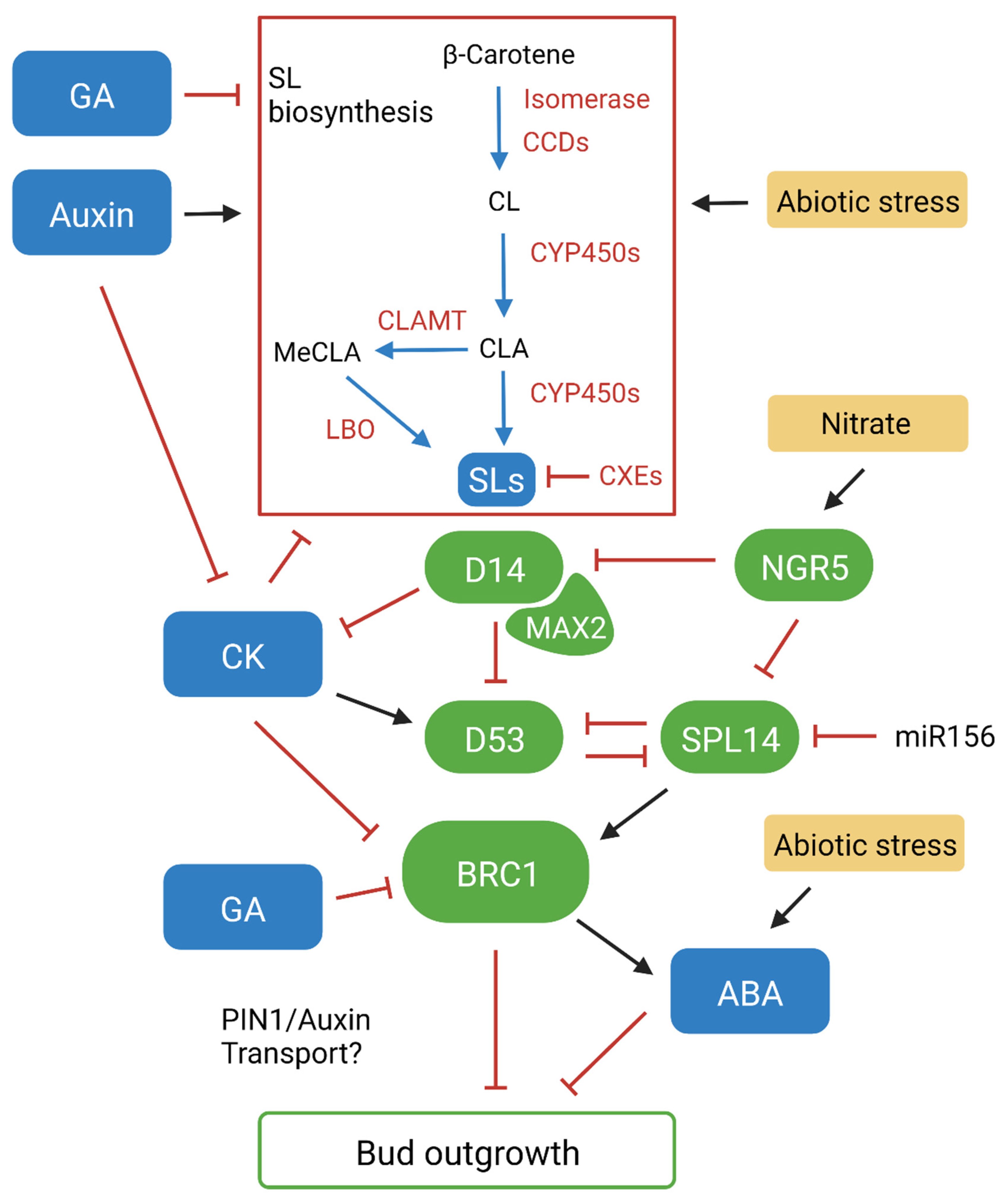
Figure 1. A complex signalling network influences branching decisions by promoting or repressing bud outgrowth. BRANCHED1 (BRC1) plays a central role within this network, acting within buds to repress outgrowth. Auxin and SL act as inducers of BRC1 while cytokinin (CK) and gibberellin (GA) act to repress it. Abscisic acid (ABA) can also act as a negative regulator of bud outgrowth downstream of BRC1. Blue arrow, conversion; black arrow, promotion effect; red line, inhibition effect; green element, proteins/transcription factors; blue element, phytohormones; yellow element, abiotic condition. Created with BioRender.com.
3. Strigolactone Signalling Mechanism
The α/β-hydrolase DWARF 14 (D14) was initially identified as the receptor for SLs in rice (Oryza sativa) tillering mutants and has since been isolated in numerous species including Arabidopsis (AtD14), petunia (Petunia hybrida) (DAD2) and barley (Hordeum vulgare) (HvD14) (Figure 1) [24][25][26][27]. D14 forms the core of SL signal perception, where it can directly bind SL molecules. The binding of an SL promotes the interaction between D14 and an F-box protein originally identified in Arabidopsis as MORE AXILLARY GROWTH 2 (MAX2), forming a SKP1-CULLIN-F-BOX (SCF) ubiquitin ligase complex [28]. Observations of max2 mutants showed the same high tillering phenotype as SL biosynthesis mutants but could not be rescued with treatment of synthetic SL (GR24), highlighting that SL signalling is dependent on D14-MAX2 for proteasome-mediated protein degradation [29]. The target proteins were identified as transcriptional repressors (represented by DWARF 53 (D53) in rice) [30][31][32]. After elucidating the function of these proteins, the general mode of action for SL signal transduction could be proposed. SL binds to D14, which then recruits the MAX2 F-box protein and D53 target proteins to form an SCF complex. D53 then undergoes proteasomal degradation, triggering the downstream SL signalling response [33].
The D14 receptor is somewhat unique compared to other hormone receptors, due to its dual function as a receptor and an enzyme. When SL is bound, the ABC-ring is cleaved from the D-ring, releasing the ABC-ring from D14 resulting in the creation of a ‘covalently linked intermediate molecule’ (CLIM) [34]. It was proposed that the creation of CLIM allows for the conformational change of D14, allowing for interaction with key proteins, such as D53 [35]. However, subsequent findings found that the D-ring may be released as a product of the reaction rather than being bound as CLIM, and that conformational changes in the α-helix of the F-box protein determine D14 conformation [36]. While modelling of the SL signal transduction mechanism is still ongoing, these findings propose that upon binding of SL, the conformation of the α-helix in the F-box protein changes the conformation of D14, which determines if the entire SL molecule is bound, or if the D-ring is cleaved at the enol bridge to regulate SL activity. After D14 conformation change it can interact with the SCF complex and recruit target proteins, where degradation and ubiquitination can then occur to trigger a response [35]. These findings underline the unique properties and significance of D14 in regulating SL signal perception, highlighting it as a key component of SL-mediated growth response. D14 seems to have evolved only in seed plants, perhaps from the receptor of the karrikin pathway, with which it retains some cross-functionality [37]. Although SLs can trigger responses in non-seed plants and microorganisms, the SL receptor in other species remains unknown.
4. Strigolactone-Mediated Bud Outgrowth
The involvement of SLs in the regulation of shoot architecture has been extensively investigated since the initial discovery of the shoot multiplication signal (SMS) and the subsequent classification of SLs in high-branching mutants [29]. SLs were first identified to inhibit bud outgrowth in experiments including highly branched ccd8 (SL biosynthesis) and max2 (SL signalling) mutants, where it was observed that application of GR24 to buds could rescue ccd8 branching to wildtype (WT) levels, while having no effect on max2 [29][38]. It is known that bud outgrowth is regulated by a highly complex network of hormonal signals, including auxin, cytokinins (CKs), gibberellins (GAs), abscisic acid (ABA) and sucrose. Additionally, there are other effects of SLs that could impact on plant growth, such as root architecture and soil microbe symbiosis, as reviewed in [39][40]. Auxin is a key growth hormone that is synthesised in shoot tips, where it then moves rootward via the polar auxin transport system (PATS) [41]. Auxin’s involvement in the regulation of bud outgrowth has been extensively investigated since its discovery by Thimann and Skoog, who showed that removal of the shoot apex in broad bean (Vicia faba) stimulated outgrowth of axillary buds, and that application of exogenous auxin to decapitated stumps could repress bud outgrowth [42]. Apical dominance is a longstanding model for auxin-mediated bud repression that has continuously evolved over time. It was initially proposed that auxin synthesised in the shoot apex moves downward into buds to inhibit them directly, although this has since been refuted as auxin from the shoot apex does not enter axillary buds in appreciable quantities, suggesting that it regulates outgrowth indirectly [43]. The auxin canalisation model proposes that auxin forms narrow transport streams that connect auxin synthesising tissues (source) to regions where auxin is being depleted (sink) [2]. Polar auxin transport occurs via the PIN-FORMED (PIN) protein efflux carrier proteins, with PIN1 being integral for facilitating downward auxin flow within the stem [44]. As part of a feedback system, auxin can promote expression of PIN genes and localise PINs facing the sink within the plasma membrane to alter the sink strength in the stem [2]. By modulating the sink strength canalisation can be promoted or repressed, determining if an axillary bud grows out into a branch. This also outlines the effect of competitive inhibition, where auxin export from a more mature bud can reduce the sink strength and prevent canalisation from younger buds, allowing it to develop into a branch while other buds remain repressed [45]. Although this informs researchers that canalisation is a necessary condition for bud outgrowth, experiments have shown that initial outgrowth can still occur in pea (Pisum sativum) plants treated with auxin transport inhibitors, suggesting that auxin canalisation is more important for ongoing bud outgrowth, rather than initiation [46].
The interaction between auxin and SL was first identified in Arabidopsis SL biosynthesis mutants which showed elevated levels of PIN1 [47]. Subsequent findings also identified a promotive effect of auxin on SL biosynthesis, and that GR24 only inhibited bud outgrowth in the presence of auxin in the main stem [48]. This highlights a homeostatic feedback loop between auxin and SL and suggests that SLs play a key role in the auxin canalisation model, where they are transported upward to repress bud outgrowth via modulating PIN1 levels to promote or repress auxin export from axillary buds. While this infers that SL-mediated bud repression is auxin dependant, it has also been identified that SLs can act downstream of auxin signalling to repress bud outgrowth. Experiments conducted in pea found that applying GR24 could inhibit bud outgrowth, even when auxin was depleted in the stem following decapitation [49]. It has also been observed that application of GR24 to shoots treated with the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) can still inhibit bud outgrowth, suggesting that SL can repress branching independently of auxin [50].
This is further supported by the identification of the BRANCHED1 (BRC1) transcription factor. BRC1 expression is highly localised in developing buds and has been observed to arrest their outgrowth, keeping them in a state of dormancy [51]. Like SL mutants, brc1 mutants exhibit a high branching phenotype which cannot be rescued with GR24, suggesting that BRC1 functions downstream of SL [49]. BRC1 expression is also reduced in SL mutants and has been observed to be upregulated by GR24 in pea [52]. This highlights that BRC1 acts as an integrator in SL-mediated branching responses, where auxin promotes SL expression, which subsequently promotes BRC1 expression in buds to inhibit outgrowth (Figure 1). While SLs act to induce BRC1 expression inside buds, it has been shown that CK acts antagonistically to repress it [52]. In contrast to SLs, auxin is known to downregulate CK levels, which has been shown to subsequently downregulate BRC1 to promote bud outgrowth in pea [53]. Experiments in Arabidopsis have also shown that CK can regulate lateral auxin transport by promoting PIN3,4,7 accumulation, suggesting that it can also influence auxin canalisation independently of BRC1 [54]. GA is another positive regulator of growth that has been linked to branching, with observations in rice showing that GAs regulate SL biosynthesis, and in Rosa sp. showing that GA biosynthesis is strongly upregulated in buds during outgrowth [55][56]. GA can also function synergistically with CK to negatively regulate BRC1 and promote bud outgrowth in Jatropha curcas [57]. These findings propose that BRC1 is a central regulator of branching that is modulated by the upstream regulation of SL, CK and GA (Figure 1). Experiments in Arabidopsis have shown that ABA levels decrease in correlation with dormancy release, and that expression is upregulated in wildtype plants treated with red/far red light, but not in brc1 mutants [58]. These results suggest that ABA can also regulate bud outgrowth via downstream repression of BRC1-mediated branching. The involvement and interaction between these hormones highlights that bud outgrowth is regulated via a highly complex signalling network, where multiple hormonal pathways can promote and repress lateral branching by manipulation of auxin transport, or by independently regulating BRC1-mediated branching [59]. This network forms the basis of the second messenger model for apical dominance, which suggests that apically derived auxin interacts with and modulates other key phytohormones to regulate bud outgrowth. While the proposed models for apical dominance and bud repression continue to evolve, SLs play an essential role in the signalling responses that facilitate important branching decisions.
References
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236.
- Zhang, J.; Mazur, E.; Balla, J.; Gallei, M.; Kalousek, P.; Medvedova, Z.; Li, Y.; Wang, Y.; Prat, T.; Vasileva, M.; et al. Strigolactones inhibit auxin feedback on PIN-dependent auxin transport canalization. Nat. Commun. 2020, 11, 3508.
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 1966, 154, 1189–1190.
- Yoneyama, K.; Brewer, P.B. Strigolactones, how are they synthesized to regulate plant growth and development? Curr. Opin. Plant Biol. 2021, 63, 102072.
- Lopez-Obando, M.; Ligerot, Y.; Bonhomme, S.; Boyer, F.D.; Rameau, C. Strigolactone biosynthesis and signaling in plant development. Development 2015, 142, 3615–3619.
- Yokota, T.; Sakai, H.; Okuno, K.; Yoneyama, K.; Takeuchi, Y. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 1998, 49, 1967–1973.
- Zwanenburg, B.; Pospisil, T. Structure and activity of strigolactones: New plant hormones with a rich future. Mol. Plant 2013, 6, 38–62.
- Yoneyama, K. Recent progress in the chemistry and biochemistry of strigolactones. J. Pestic. Sci. 2020, 45, 45–53.
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351.
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238.
- Sorefan, K.; Booker, J.; Haurogne, K.; Goussot, M.; Bainbridge, K.; Foo, E.; Chatfield, S.; Ward, S.; Beveridge, C.; Rameau, C.; et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474.
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645.
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449.
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089.
- Yoneyama, K.; Mori, N.; Sato, T.; Yoda, A.; Xie, X.; Okamoto, M.; Iwanaga, M.; Ohnishi, T.; Nishiwaki, H.; Asami, T.; et al. Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 2018, 218, 1522–1533.
- Xie, X.; Yoneyama, K.; Kisugi, T.; Uchida, K.; Ito, S.; Akiyama, K.; Hayashi, H.; Yokota, T.; Nomura, T.; Yoneyama, K. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 2013, 6, 153–163.
- Mashiguchi, K.; Seto, Y.; Onozuka, Y.; Suzuki, S.; Takemoto, K.; Wang, Y.; Dong, L.; Asami, K.; Noda, R.; Kisugi, T.; et al. A carlactonoic acid methyltransferase that contributes to the inhibition of shoot branching in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2111565119.
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306.
- Xu, E.; Chai, L.; Zhang, S.; Yu, R.; Zhang, X.; Xu, C.; Hu, Y. Catabolism of strigolactones by a carboxylesterase. Nat. Plants 2021, 7, 1495–1504.
- Roesler, K.; Lu, C.; Thomas, J.; Xu, Q.; Vance, P.; Hou, Z.; Williams, R.W.; Liu, L.; Owens, M.A.; Habben, J.E. Arabidopsis Carboxylesterase 20 Binds Strigolactone and Increases Branches and Tillers When Ectopically Expressed in Arabidopsis and Maize. Front. Plant Sci. 2021, 12, 639401.
- Marzec, M.; Situmorang, A.; Brewer, P.B.; Braszewska, A. Diverse Roles of MAX1 Homologues in Rice. Genes 2020, 11, 1348.
- Shiratake, K.; Notaguchi, M.; Makino, H.; Sawai, Y.; Borghi, L. Petunia PLEIOTROPIC DRUG RESISTANCE 1 is a Strigolactone Short-Distance Transporter with Long-Distance Outcomes. Plant Cell Physiol. 2019, 60, 1722–1733.
- Ito, S.; Braguy, J.; Wang, J.Y.; Yoda, A.; Fiorilli, V.; Takahashi, I.; Jamil, M.; Felemban, A.; Miyazaki, S.; Mazzarella, T.; et al. Canonical strigolactones are not the major determinant of tillering but important rhizospheric signals in rice. Sci. Adv. 2022, 8, eadd1278.
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424.
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295.
- Hamiaux, C.; Drummond, R.S.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036.
- Marzec, M.; Gruszka, D.; Tylec, P.; Szarejko, I. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant 2016, 158, 341–355.
- Stirnberg, P.; Furner, I.J.; Ottoline Leyser, H.M. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007, 50, 80–94.
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194.
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410.
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405.
- Wang, L.; Wang, B.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G.; et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281.
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322.
- Yao, R.; Ming, Z.; Yan, L.; Li, S.; Wang, F.; Ma, S.; Yu, C.; Yang, M.; Chen, L.; Chen, L.; et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016, 536, 469–473.
- Marzec, M.; Brewer, P. Binding or Hydrolysis? How Does the Strigolactone Receptor Work? Trends Plant Sci. 2019, 24, 571–574.
- Shabek, N.; Ticchiarelli, F.; Mao, H.; Hinds, T.R.; Leyser, O.; Zheng, N. Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 2018, 563, 652–656.
- Li, Q.; Martin-Fontecha, E.S.; Khosla, A.; White, A.R.F.; Chang, S.; Cubas, P.; Nelson, D.C. The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Commun. 2022, 3, 100303.
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200.
- Sun, H.; Li, W.; Burritt, D.J.; Tian, H.; Zhang, H.; Liang, X.; Miao, Y.; Mostofa, M.G.; Tran, L.-S.P. Strigolactones interact with other phytohormones to modulate plant root growth and development. Crop J. 2022, 10, 1517–1527.
- Kaniganti, S.; Bhattacharya, J.; Petla, B.P.; Reddy, P.S. Strigolactone, a neglected plant hormone, with a great potential for crop improvement: Crosstalk with other plant hormones. Environ. Exp. Bot. 2022, 204, 105072.
- Bennett, T.; Hines, G.; van Rongen, M.; Waldie, T.; Sawchuk, M.G.; Scarpella, E.; Ljung, K.; Leyser, O. Connective Auxin Transport in the Shoot Facilitates Communication between Shoot Apices. PLoS Biol. 2016, 14, e1002446.
- Thimann, K.V.; Skoog, F. Studies on the Growth Hormone of Plants: III. The Inhibiting Action of the Growth Substance on Bud Development. Proc. Natl. Acad. Sci. USA 1933, 19, 714–716.
- Hall, S.M.; Hillman, J.R. Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. timing of bud growth following decapitation. Planta 1975, 123, 137–143.
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883.
- Balla, J.; Medvedova, Z.; Kalousek, P.; Matijescukova, N.; Friml, J.; Reinohl, V.; Prochazka, S. Auxin flow-mediated competition between axillary buds to restore apical dominance. Sci. Rep. 2016, 6, 35955.
- Chabikwa, T.G.; Brewer, P.B.; Beveridge, C.A. Initial Bud Outgrowth Occurs Independent of Auxin Flow from Out of Buds. Plant Physiol. 2019, 179, 55–65.
- Bennett, T.; Sieberer, T.; Willett, B.; Booker, J.; Luschnig, C.; Leyser, O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 2006, 16, 553–563.
- Crawford, S.; Shinohara, N.; Sieberer, T.; Williamson, L.; George, G.; Hepworth, J.; Muller, D.; Domagalska, M.A.; Leyser, O. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 2010, 137, 2905–2913.
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009, 150, 482–493.
- Brewer, P.B.; Dun, E.A.; Gui, R.; Mason, M.G.; Beveridge, C.A. Strigolactone Inhibition of Branching Independent of Polar Auxin Transport. Plant Physiol. 2015, 168, 1820–1829.
- Aguilar-Martinez, J.A.; Poza-Carrion, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472.
- Dun, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012, 158, 487–498.
- Braun, N.; de Saint Germain, A.; Pillot, J.P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238.
- Waldie, T.; Leyser, O. Cytokinin Targets Auxin Transport to Promote Shoot Branching. Plant Physiol. 2018, 177, 803–818.
- Choubane, D.; Rabot, A.; Mortreau, E.; Legourrierec, J.; Peron, T.; Foucher, F.; Ahcene, Y.; Pelleschi-Travier, S.; Leduc, N.; Hamama, L.; et al. Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J. Plant Physiol. 2012, 169, 1271–1280.
- Ito, S.; Yamagami, D.; Umehara, M.; Hanada, A.; Yoshida, S.; Sasaki, Y.; Yajima, S.; Kyozuka, J.; Ueguchi-Tanaka, M.; Matsuoka, M.; et al. Regulation of Strigolactone Biosynthesis by Gibberellin Signaling. Plant Physiol. 2017, 174, 1250–1259.
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin Promotes Shoot Branching in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666.
- Gonzalez-Grandio, E.; Poza-Carrion, C.; Sorzano, C.O.; Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 2013, 25, 834–850.
- Wang, M.; Le Moigne, M.A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.D.; Oge, L.; Demotes-Mainard, S.; Hamama, L.; Daviere, J.M.; Sakr, S. BRANCHED1: A Key Hub of Shoot Branching. Front. Plant Sci. 2019, 10, 76.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
3 times
(View History)
Update Date:
17 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




