Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Masaya Baba | -- | 1085 | 2023-01-16 08:54:06 | | | |
| 2 | Conner Chen | + 3 word(s) | 1088 | 2023-01-16 09:55:08 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tang, J.; Baba, M. Microphthalmia Family Translocation Renal Cell Carcinoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/40199 (accessed on 07 February 2026).
Tang J, Baba M. Microphthalmia Family Translocation Renal Cell Carcinoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/40199. Accessed February 07, 2026.
Tang, Jinglong, Masaya Baba. "Microphthalmia Family Translocation Renal Cell Carcinoma" Encyclopedia, https://encyclopedia.pub/entry/40199 (accessed February 07, 2026).
Tang, J., & Baba, M. (2023, January 16). Microphthalmia Family Translocation Renal Cell Carcinoma. In Encyclopedia. https://encyclopedia.pub/entry/40199
Tang, Jinglong and Masaya Baba. "Microphthalmia Family Translocation Renal Cell Carcinoma." Encyclopedia. Web. 16 January, 2023.
Copy Citation
The microphthalmia-associated transcription factor/transcription factor E (MiT/TFE) family of transcription factors are evolutionarily conserved, basic helix–loop–helix leucine zipper (bHLH-Zip) transcription factors, consisting of MITF, TFEB, TFE3, and TFEC. MiT/TFE proteins, with the exception of TFEC, are involved in the development of renal cell carcinoma (RCC). Most of the MiT/TFE transcription factor alterations seen in sporadic RCC cases of MiT family translocation renal cell carcinoma (tRCC) are chimeric proteins generated by chromosomal rearrangements. These chimeric MiT/TFE proteins retain the bHLH-Zip structures and act as oncogenic transcription factors.
MiT/TFE family
RCC
MITF
1. The Microphthalmia/Transcription Factor E (MiT/TFE) Family
The microphthalmia-associated transcription factor/transcription factor E (MiT/TFE) family consists of MITF, TFEB, TFE3, and TFEC. All of the proteins have a bHLH-Zip (basic helix–loop–helix leucine zipper) structure, which allows them to form homodimers or heterodimers with each other and bind to the regulatory elements of target genes [1][2][3][4]. The binding consensus sequences recognized by the MiT/TFE proteins are known as the E-box motif (CACGTG) and M-box motif (TCATGTG) [5][6][7][8]. The MiT/TFE proteins are the master regulators of lysosomal biogenesis and autophagy [9]. In addition, their fundamental roles in many cellular processes including proliferation, differentiation, survival, senescence, invasion, metabolism, organelle biogenesis, and stress responses are emerging [1][2][3]. Both MiT family translocation renal cell carcinoma (tRCC) and MITF p.E318K renal cell carcinoma (RCC) are rare and not widely recognized by clinicians. In the future, it will be necessary to increase awareness of these MiT/TFE family RCCs and develop biomarkers to facilitate diagnosis.
2. MiT Family Translocation Renal Cell Carcinoma (tRCC)
MiT family translocation renal cell carcinoma (tRCC) is a sporadic RCC characterized by fusion genes involving the MiT/TFE family genes, MITF, TFEB, and TFE3 and defined as an MiT family translocation RCC in the 2016 WHO classification [10][11]. In the 2022 WHO classification, tRCC is divided into TFE3-rearranged RCC and TFEB-altered RCC as Moleculary-defined RCCs [12]. tRCC is a rare disease that accounts for approximately 1–5% of sporadic RCC in adults [13][14][15][16][17], developing more often in women than in men, and is much more commonly seen in pediatric RCC cases (approximately 40% (range 20–75%)) [13][18][19][20][21]. tRCC tends to be in an advanced stage at onset with a more aggressive presentation than other sporadic RCCs, and molecular targeted therapy for advanced cases has not yet been established [10][22]. The most distinctive histopathological features of tRCC are clear-cell papillary, displaying a papillary structure consisting of clear cells. However, recent studies have shown that tRCCs are morphologically heterogenous and can include papillary, tubular, acinar, and even cystic architecture [10][17][23][24]. In fact, some cases are indistinguishable from clear-cell (cc)RCC or papillary (p)RCC by H&E staining alone [16]. Therefore, it is expected that the actual number of tRCC cases may be higher than the number currently diagnosed. Definitive diagnosis requires TFE3/TFEB/MITF immunohistochemistry and FISH [25][26][27], which are not routinely performed in many hospitals. Hence, the immunohistochemistry of surrogate markers such as cathepsin K, Melan A, and GPNMB should be considered for the initial diagnosis of RCC [18][24][28]. Chimeric MiT/TFE proteins are generated by chromosomal rearrangements. Bakouny et al. have reported that among 88 fusion-defined tRCC cases, most fusion genes (88.6%) involved TFE3. On the other hand, TFEB fusions were reported in only 9.1% and MITF fusions in only 2.3% of these cases [16]. A single center study of the largest TFE3-rearranged RCC cohort published to date identified 57 TFE3 fusion genes in 4581 RCCs [17]. Of the 57 cases, 26.3% were SFPQ-TFE3 fusions and 22.8% were ASPSCR1-TFE3 fusions. Of note, X chromosomal inversion was noted in 21% (12/57) of cases, which consisted of NONO-TFE3 (14%) and RBM10-TFE3 (7%). These X chromosomal inversion cases can be misdiagnosed as non-TFE3-rearranged RCC because of the narrow interval between split signals seen with TFE3 gene break-apart FISH, the gold standard diagnostic test for TFE3-rearranged RCC [29][30]. (Figure 1a–d) TFEB-altered RCC includes 6p21.1 translocated RCC and 6p21.1 amplified RCC, which demonstrate distinct signals seen with TFEB gene break-apart FISH (Figure 1e,f) [31]. All MiT/TFE fusion genes identified to date retain the bHLH-Zip structure (Figure 2) [16][17][22][32][33][34][35][36], suggesting that these MiT/TFE fusion genes function as oncogenic transcription factors. Indeed, overexpression of TFE3 fusion, PRCC–TFE3, in mouse kidneys was shown to cause RCC with aberrant expression of MiT/TFE target genes [28]. There are few recurrent genomic alterations in tRCC other than MiT/TFE gene rearrangement and 9p21.3 deletion [16]. Several genomic alterations, such as ASPSCR1-TFE3, LUC7L3-TFE3, and 22q deletion, correlate with poor prognosis [17][18]. Currently, there is no established standard therapy for advanced tRCC [10][22]. However, recent studies suggest that immunotherapy may be effective for advanced tRCC [16][17].
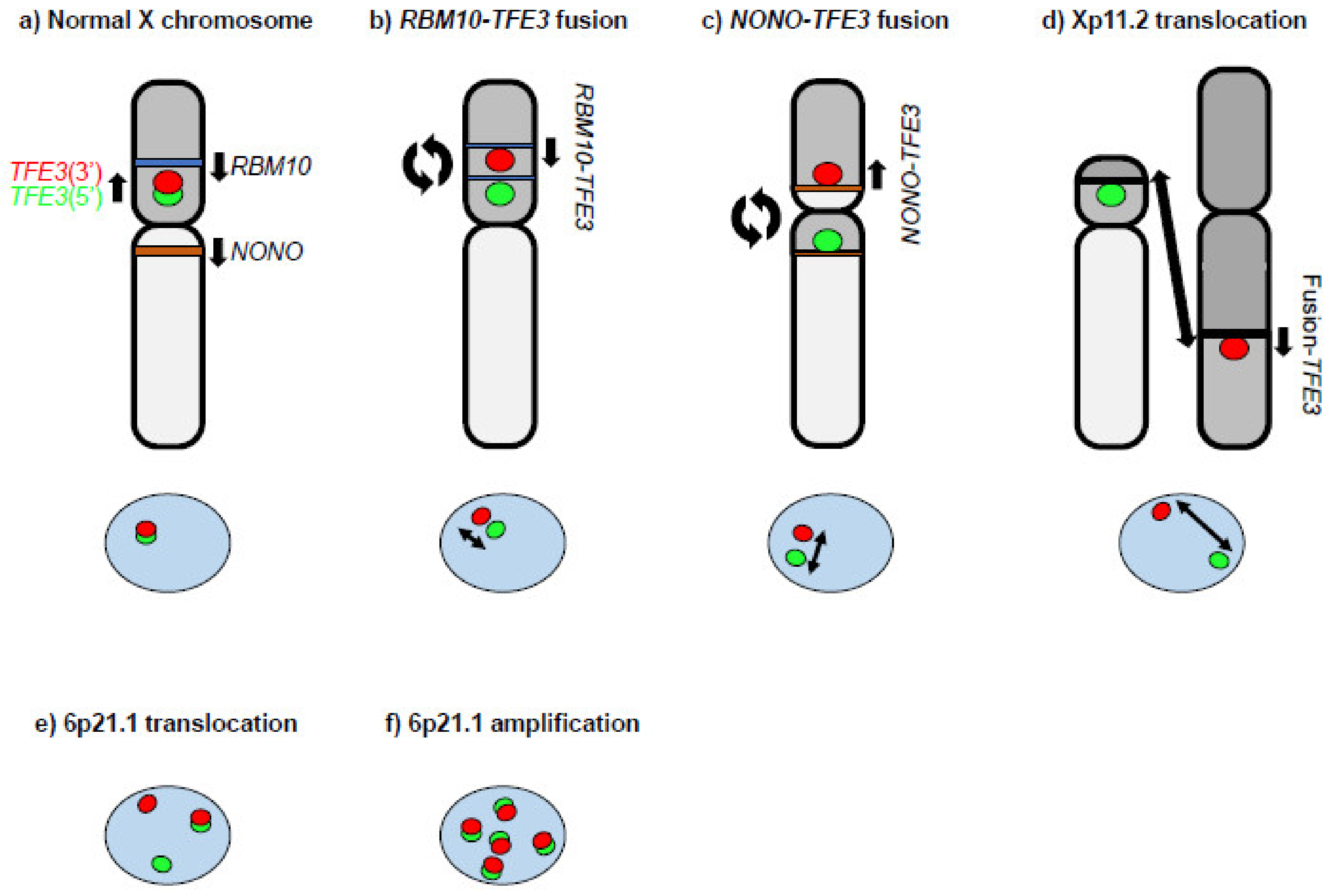
Figure 1. A diagram of TFE3 gene break-apart FISH. (a) The TFE3 gene is located at chromosome Xp11.2. The 5′ FISH probe for TFE3 is green. The 3′ FISH probe for TFE3 is red. A fusion candidate gene, RBM10 (blue bar), is located at chromosome Xp11.23 on the telomere side of TFE3. Another fusion candidate gene, NONO (brown bar), is located at chromosome Xq13.1. The TFE3 gene break-apart FISH demonstrates co-localization of the green and red probes. (b) An intra Xp (paracentric) inversion inv(X)(p11.2;p11.23) causes the RBM10–TFE3 fusion gene. The TFE3 gene break-apart FISH demonstrates subtle split red and green signals. Two blue bars indicate separated RBM10. The length of the two-arrowhead line indicates the relative distance between the FISH signals. (c) A pericentric X chromosome inversion, inv(X)(p11.2;q13.1), causes the NONO–TFE3 fusion gene. The TFE3 gene break-apart FISH demonstrates slight split red and green signals. The two brown bars indicate separated NONO. The length of the two-arrowhead line indicates the relative distance between the FISH signals. (d) Xp11.2 translocation may occur with another chromosome. The TFE3 gene break-apart FISH demonstrates clearly separated green and red signals. The length of the two-arrowhead line indicates the relative distance between the FISH signals. (e) The TFEB gene is located at chromosome 6p21.1. The 5′ FISH probe for TFEB is green. The 3′ FISH probe for TFEB is red. 6p21.1 translocated RCC demonstrates clearly separated green and red signals as well as co-localized green and red signals by TFEB gene break-apart FISH. (f) 6p21.1 amplified RCC demonstrates amplification of co-localized green and red signals by TFEB gene break-apart FISH.

Figure 2. Structures of TFE3 fusion and TFEB fusion genes. The structures of TFE3 fusions and TFEB fusions found in TFE3-rearranged RCC and TFEB-altered RCC are shown. Wild-type TFE3 and TFEB have 10 exons. The fusion partner genes are listed. All of the fusion genes retain coding exons for the bHLH-Zip domain. AD: activation domain; bHLH-Zip: basic helix–loop–helix leucine zipper.
References
- Goding, C.R.; Arnheiter, H. MITF-the first 25 years. Genes Dev. 2019, 33, 983–1007.
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 129, 2475–2481.
- La Spina, M.; Contreras, P.S.; Rissone, A.; Meena, N.K.; Jeong, E.; Martina, J.A. MiT/TFE Family of Transcription Factors: An Evolutionary Perspective. Front. Cell Dev. Biol. 2020, 8, 609683.
- Beckmann, H.; Kadesch, T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991, 5, 1057–1066.
- Hemesath, T.J.; Steingrimsson, E.; McGill, G.; Hansen, M.J.; Vaught, J.; Hodgkinson, C.A.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A.; Fisher, D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994, 8, 2770–2780.
- Strub, T.; Giuliano, S.; Ye, T.; Bonet, C.; Keime, C.; Kobi, D.; Le Gras, S.; Cormont, M.; Ballotti, R.; Bertolotto, C.; et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 2011, 30, 2319–2332.
- Aksan, I.; Goding, C.R. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 1998, 18, 6930–6938.
- Pogenberg, V.; Ogmundsdottir, M.H.; Bergsteinsdottir, K.; Schepsky, A.; Phung, B.; Deineko, V.; Milewski, M.; Steingrimsson, E.; Wilmanns, M. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012, 26, 2647–2658.
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433.
- Argani, P. Translocation carcinomas of the kidney. Genes Chromosomes Cancer 2022, 61, 219–227.
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105.
- Moch, H.; Amin, M.B.; Berney, D.M.; Comperat, E.M.; Gill, A.J.; Hartmann, A.; Menon, S.; Raspollini, M.R.; Rubin, M.A.; Srigley, J.R.; et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2022, 82, 458–468.
- Sukov, W.R.; Hodge, J.C.; Lohse, C.M.; Leibovich, B.C.; Thompson, R.H.; Pearce, K.E.; Wiktor, A.E.; Cheville, J.C. TFE3 rearrangements in adult renal cell carcinoma: Clinical and pathologic features with outcome in a large series of consecutively treated patients. Am. J. Surg. Pathol. 2012, 36, 663–670.
- Komai, Y.; Fujiwara, M.; Fujii, Y.; Mukai, H.; Yonese, J.; Kawakami, S.; Yamamoto, S.; Migita, T.; Ishikawa, Y.; Kurata, M.; et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin. Cancer Res. 2009, 15, 1170–1176.
- Zhong, M.; De Angelo, P.; Osborne, L.; Paniz-Mondolfi, A.E.; Geller, M.; Yang, Y.; Linehan, W.M.; Merino, M.J.; Cordon-Cardo, C.; Cai, D. Translocation renal cell carcinomas in adults: A single-institution experience. Am. J. Surg. Pathol. 2012, 36, 654–662.
- Bakouny, Z.; Sadagopan, A.; Ravi, P.; Metaferia, N.Y.; Li, J.; AbuHammad, S.; Tang, S.; Denize, T.; Garner, E.R.; Gao, X.; et al. Integrative clinical and molecular characterization of translocation renal cell carcinoma. Cell Rep. 2022, 38, 110190.
- Sun, G.; Chen, J.; Liang, J.; Yin, X.; Zhang, M.; Yao, J.; He, N.; Armstrong, C.M.; Zheng, L.; Zhang, X.; et al. Integrated exome and RNA sequencing of TFE3-translocation renal cell carcinoma. Nat. Commun. 2021, 12, 5262.
- Qu, Y.; Wu, X.; Anwaier, A.; Feng, J.; Xu, W.; Pei, X.; Zhu, Y.; Liu, Y.; Bai, L.; Yang, G.; et al. Proteogenomic characterization of MiT family translocation renal cell carcinoma. Nat. Commun. 2022, 13, 7494.
- Van der Beek, J.N.; Hol, J.A.; Coulomb-l’Hermine, A.; Graf, N.; van Tinteren, H.; Pritchard-Jones, K.; Houwing, M.E.; de Krijger, R.R.; Vujanic, G.M.; Dzhuma, K.; et al. Characteristics and outcome of pediatric renal cell carcinoma patients registered in the International Society of Pediatric Oncology (SIOP) 93-01, 2001 and UK-IMPORT database: A report of the SIOP-Renal Tumor Study Group. Int. J. Cancer 2021, 148, 2724–2735.
- Van der Beek, J.N.; Geller, J.I.; de Krijger, R.R.; Graf, N.; Pritchard-Jones, K.; Drost, J.; Verschuur, A.C.; Murphy, D.; Ray, S.; Spreafico, F.; et al. Characteristics and Outcome of Children with Renal Cell Carcinoma: A Narrative Review. Cancers 2020, 12, 1776.
- Argani, P. MiT family translocation renal cell carcinoma. Semin. Diagn. Pathol. 2015, 32, 103–113.
- Kauffman, E.C.; Ricketts, C.J.; Rais-Bahrami, S.; Yang, Y.; Merino, M.J.; Bottaro, D.P.; Srinivasan, R.; Linehan, W.M. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat. Rev. Urol. 2014, 11, 465–475.
- Kuroda, N.; Mikami, S.; Pan, C.C.; Cohen, R.J.; Hes, O.; Michal, M.; Nagashima, Y.; Tanaka, Y.; Inoue, K.; Shuin, T.; et al. Review of renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions with focus on pathobiological aspect. Histol. Histopathol. 2012, 27, 133–140.
- Caliò, A.; Brunelli, M.; Segala, D.; Pedron, S.; Remo, A.; Ammendola, S.; Munari, E.; Pierconti, F.; Mosca, A.; Bollito, E.; et al. Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: Investigating prognostic markers and therapy targets. Pathology 2020, 52, 297–309.
- Zhong, M.; De Angelo, P.; Osborne, L.; Keane-Tarchichi, M.; Goldfischer, M.; Edelmann, L.; Yang, Y.; Linehan, W.M.; Merino, M.J.; Aisner, S.; et al. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am. J. Surg. Pathol. 2010, 34, 757–766.
- Argani, P.; Yonescu, R.; Morsberger, L.; Morris, K.; Netto, G.J.; Smith, N.; Gonzalez, N.; Illei, P.B.; Ladanyi, M.; Griffin, C.A. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am. J. Surg. Pathol. 2012, 36, 1516–1526.
- Skala, S.L.; Xiao, H.; Udager, A.M.; Dhanasekaran, S.M.; Shukla, S.; Zhang, Y.; Landau, C.; Shao, L.; Roulston, D.; Wang, L.; et al. Detection of 6 TFEB-amplified renal cell carcinomas and 25 renal cell carcinomas with MITF translocations: Systematic morphologic analysis of 85 cases evaluated by clinical TFE3 and TFEB FISH assays. Mod. Pathol. 2018, 31, 179–197.
- Baba, M.; Furuya, M.; Motoshima, T.; Lang, M.; Funasaki, S.; Ma, W.; Sun, H.W.; Hasumi, H.; Huang, Y.; Kato, I.; et al. TFE3 Xp11.2 Translocation Renal Cell Carcinoma Mouse Model Reveals Novel Therapeutic Targets and Identifies GPNMB as a Diagnostic Marker for Human Disease. Mol. Cancer Res. 2019, 17, 1613–1626.
- Kato, I.; Furuya, M.; Baba, M.; Kameda, Y.; Yasuda, M.; Nishimoto, K.; Oyama, M.; Yamasaki, T.; Ogawa, O.; Niino, H.; et al. RBM10-TFE3 renal cell carcinoma characterised by paracentric inversion with consistent closely split signals in break-apart fluorescence in-situ hybridisation: Study of 10 cases and a literature review. Histopathology 2019, 75, 254–265.
- Liu, N.; Guo, W.; Shi, Q.; Zhuang, W.; Pu, X.; Chen, S.; Qu, F.; Xu, L.; Zhao, X.; Li, X.; et al. The suitability of NONO-TFE3 dual-fusion FISH assay as a diagnostic tool for NONO-TFE3 renal cell carcinoma. Sci. Rep. 2020, 10, 16361.
- Gupta, S.; Argani, P.; Jungbluth, A.A.; Chen, Y.B.; Tickoo, S.K.; Fine, S.W.; Gopalan, A.; Al-Ahmadie, H.A.; Sirintrapun, S.J.; Sanchez, A.; et al. TFEB Expression Profiling in Renal Cell Carcinomas: Clinicopathologic Correlations. Am. J. Surg. Pathol. 2019, 43, 1445–1461.
- Xia, Q.Y.; Wang, X.T.; Fang, R.; Wang, Z.; Zhao, M.; Chen, H.; Chen, N.; Teng, X.D.; Wang, X.; Wei, X.; et al. Clinicopathologic and Molecular Analysis of the TFEB Fusion Variant Reveals New Members of TFEB Translocation Renal Cell Carcinomas (RCCs): Expanding the Genomic Spectrum. Am. J. Surg. Pathol. 2020, 44, 477–489.
- Antic, T.; Taxy, J.B.; Alikhan, M.; Segal, J. Melanotic Translocation Renal Cell Carcinoma With a Novel ARID1B-TFE3 Gene Fusion. Am. J. Surg. Pathol. 2017, 41, 1576–1580.
- Huang, W.; Goldfischer, M.; Babyeva, S.; Mao, Y.; Volyanskyy, K.; Dimitrova, N.; Fallon, J.T.; Zhong, M. Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer 2015, 54, 500–505.
- Malouf, G.G.; Su, X.; Yao, H.; Gao, J.; Xiong, L.; He, Q.; Comperat, E.; Couturier, J.; Molinie, V.; Escudier, B.; et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin. Cancer Res. 2014, 20, 4129–4140.
- Wei, S.; Testa, J.R.; Argani, P. A review of neoplasms with MITF/MiT family translocations. Histol. Histopathol. 2022, 37, 311–321.
More
Information
Subjects:
Genetics & Heredity
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
640
Revisions:
2 times
(View History)
Update Date:
16 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




