Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pierluigi Reveglia | -- | 2703 | 2023-01-13 10:48:37 | | | |
| 2 | Amina Yu | -3 word(s) | 2700 | 2023-01-16 03:11:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Reveglia, P.; Billones-Baaijens, R.; Savocchia, S. Phytotoxic Metabolites and Fungi of Grapevine Trunk Diseases. Encyclopedia. Available online: https://encyclopedia.pub/entry/40154 (accessed on 08 February 2026).
Reveglia P, Billones-Baaijens R, Savocchia S. Phytotoxic Metabolites and Fungi of Grapevine Trunk Diseases. Encyclopedia. Available at: https://encyclopedia.pub/entry/40154. Accessed February 08, 2026.
Reveglia, Pierluigi, Regina Billones-Baaijens, Sandra Savocchia. "Phytotoxic Metabolites and Fungi of Grapevine Trunk Diseases" Encyclopedia, https://encyclopedia.pub/entry/40154 (accessed February 08, 2026).
Reveglia, P., Billones-Baaijens, R., & Savocchia, S. (2023, January 13). Phytotoxic Metabolites and Fungi of Grapevine Trunk Diseases. In Encyclopedia. https://encyclopedia.pub/entry/40154
Reveglia, Pierluigi, et al. "Phytotoxic Metabolites and Fungi of Grapevine Trunk Diseases." Encyclopedia. Web. 13 January, 2023.
Copy Citation
Grapevines are one of the most economically important crops worldwide, with approximately 48% of the world’s grape production used for wine production. Fungal diseases are limiting factors to the production of wine grapes, impacting the quality of wine. Grapevine trunk diseases (GTDs), caused by one or several fungal pathogens, cause a progressive decline in vines resulting in a loss in productivity and eventual death of the vines. Internal and external GTDs symptoms sometimes take several years to appear after infection; thus, they are considered slow-progression diseases.
Vitis vinifera
GTDs
phytotoxins
omics
1. Phytotoxic Metabolites Produced by Fungi Involved in Esca Complex
Fungi involved in Esca complex disease include Phaeomoniella chlamydospora, 27 species of Phaeoacremonium, Pleurostoma richardsiae, and ten species of Cadophora [1][2][3][4][5].
Among the different Phaeoacremonium and Cadophora spp. occurring in Esca, Phaeoacremonium minimum and Cadophora luteo-olivacea are the most prevalent [1][6].
The above-mentioned fungi belong to the Ascomycota phylum and have been isolated from grapevine necrotic wood and identified by molecular techniques using actin and β-tubulin gene regions [3]. Finally, numerous basidiomycetous species have been isolated from grapevine wood showing Esca symptoms. These fungi belong to the genera Fomitiporia, Inocutis, Inonotus, Fomitiporella, Phellinus, and Stereum. However, their role in the symptomatology of the disease requires further study [2][7].
The draft genome sequence of both P. minimum and Ph. chlamydospora are available [8][9]. In both fungi, the genome was enriched in clusters associated with secondary metabolism, including those encoding polyketide synthetases (PKS), non-ribosomal peptide synthetases (NRPS), and also secreted proteins predicted to be putative plant CAZymes [10]. More recently, comparative genome analysis and high throughput transcriptome studies have been conducted on Ph. chlamydospora [11].
The first reported PMs (Figure 1) from P. minimum were the naphthalenone pentaketides derivative named scytalone (1) and isosclerone (2) together with 4-hydroxybenzaldehyde (3) identified by spectroscopic method, essentially NMR [12][13]. The metabolites were assayed on detached grapevine leaves, compound 1 caused chlorosis, rounded to irregular, interveinal, or marginal spots, while 2 caused large, coalescent chlorotic and necrotic spots followed by withering and distortion of the lamina. The most recent report of PMs isolated from the culture filtrate of P. minimum dates back to 2004 when Mansour et al. [14] investigated the neutral organic extract and the acid organic extract. The neutral organic extract yielded scytalone (1), isosclerone (2), 4-hydroscytalone (4), 2,4,8-trihydroxytetralone (5), 3,4,8-trihydroxytetralone (6), 1,3,8-trihydroxynaphthalene (7). The main compounds in the acidic extract were flaviolin (8), isolated with traces of 2-hydroxyjuglone (9). These compounds were assayed on grapevine callus and Arabidopsis thaliana. The isolated PMs were divided into tetralones, such as scytalone and isosclerone, which promote callus growth, and naphthoquinones, like 2-hydroxyjuglone and flaviolin, which inhibit growth [14]. Little information is known about the function of the naphthalenone pentaketides 1–9 in vine cells or tissues [13][15]. Their activity might be related to their oxidant property, especially their interaction with reactive oxygen species (ROS) produced by the plant during the defense response [16][17]. Thus, the presence of the typical tiger-stripe symptoms on leaves from grapevines infected with Esca may be related to physiological changes caused by toxic metabolites produced by the causal pathogens in the trunk. Herein, these changes would be a response of the vine to the disease [15]. When the PMs were isolated and identified from the pathogens involved in Esca and BD, this hypothesis was also extended to these two GTDs [15]. However, further investigations are needed to support this hypothesis and to correlate the production of PMs with foliar symptoms.
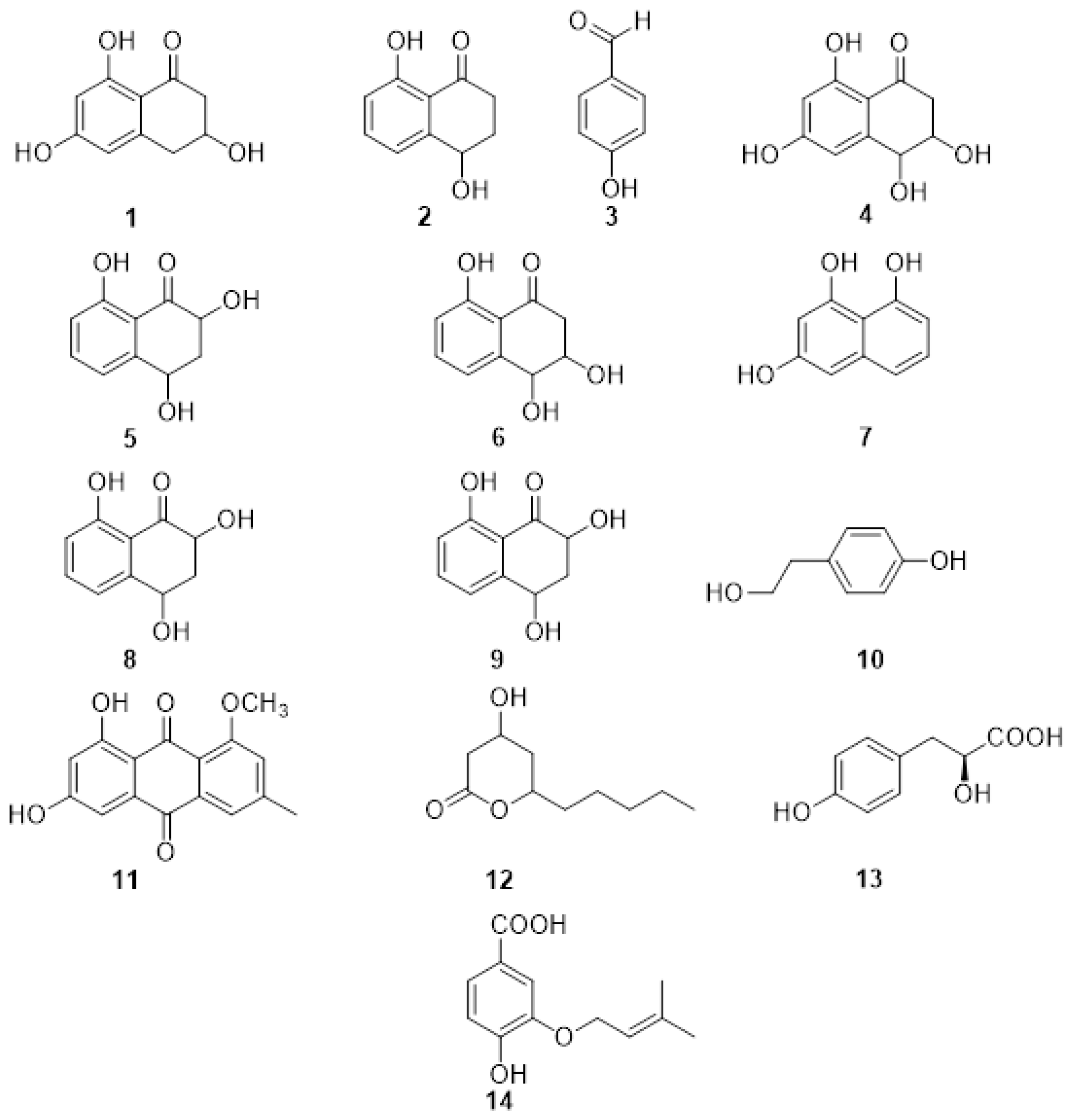
Figure 1. Structure of scytalone (1), isosclerone (2), 4-hydroxybenzaldehyde (3), 4-hydroscytalone (4), 2,4,8-trihydroxytetralone (5), 3,4,8-trihydroxytetralone (6), 1,3,8-trihydroxynaphthalene (7), flaviolin (8), 2-hydroxyjuglone (9), tyrosol (10), 1-O-methylemodine (11), 3-hydroxy-5decanolide (12), (S)-4-hydroxyphenyllactic acid (13), 3-(3-methyl-2-butenyloxy) -4-hydroxybenzoic acid (14).
Phaeomoniella chlamydospora was also investigated and produced the PMs tyrosol (10), 1-O-methylemodine (11), 3-hydroxy-5decanolide (12), (S)-4-hydroxyphenyllactic acid (13), 3-(3-methyl-2-butenyloxy)-4-hydroxybenzoic acid (14) (Figure 2). They were isolated together with the already reported scytalone (1) and isosclerone (2), and p-hydroxybenzaldehyde (3). Compound 3 showed the highest activity, inhibiting the protoplast growth of 20% at 100 mM and 100% at 1 mM. The two organic acids (S)-4-hydroxyphenyllactic acid (13), 3-(3-methyl-2-butenyloxy)-4-hydroxybenzoic acid (14) were also active on protoplasts at 1 mM concentration. Moreover, the authors point out that the aromatic aldehyde function, in the ortho or para position, is always present in the structures of the active metabolites and could be related to their activity [13].
2. Phytotoxic Metabolites Produced by Fungi Involved in Eutypa Dieback
The fungus Eutypa lata is a diatrypaceous fungus that is considered the main causal agent of Eutypa dieback. Although 22 species of diatrypaceae have been isolated from a grapevine showing typical ED symptoms [1][18][19][20], only E. lata is known to cause the typical foliar symptoms associated with this GTD [6][20][21].
These fungi have been isolated from asymptomatic tissues several centimeters ahead of lesion margins, indicating pathogen latency. Eutypa lata, among other GTD pathogens, was also found in asymptomatic grapevines aged 40 years or older, highlighting that there could be a balance between the plant microbiome and pathogenic fungi, assisting in preventing the development of the disease [22].
The first draft genome sequence of E. lata (UCR-EL1) was published in 2013 [23]. The study provided a preliminary inventory of the potential virulence factors and an abundant repertoire of cell wall-degrading enzymes, and a high number of putative cytochrome P450 monooxygenases implicated in lignin oxidation [23]. In 2015, Morales-Cruz and co-authors highlighted a great expansion of families of genes involved in the biosynthesis of toxins, including polyketide synthesis (t1PKS) [24]. More recently, the first whole-genome sequencing and comparative genomics study on 40 E. lata isolates from Australia was reported [25].
The first investigation on PMs produced by E. lata was carried out in 1989 [26], where the screening of the bioactive organic fractions led to the isolation of eight new aromatic compound metabolites characterized by a 3-methylbut-3-en-1-ynyl functional group: eutypine (15), eutypinol (16), O-methyleutypine (17), O-methyleutypinol (18), eutypune carboxylic acid (19), 4-ydroxy-3,4-dihydroxy-3-methylbut-l-ynyl)benzyl alcohol (20), 4-hydroxy-3-(3,4-dihydroxy-3-methylbut-l-ynyl)benzaldehyde (21), 3-hydroxy-3,4-diliydroxy-3-methylbut-I-ynyl)benzoic acid (22) (Figure 2). Moreover, the authors detected 5-formyl-2-(methylvinyl)[1]benzofuran (23) in the culture medium, which is also obtained from compound 1 under middle acid condition (Figure 2). The compounds were tested on tomato seedlings and grapevine leaves. Eutypine (15) showed the highest phytotoxic activity. Moreover, the authors suggested that the aldehyde group and a free OH in para-position could be necessary for the activity [27].
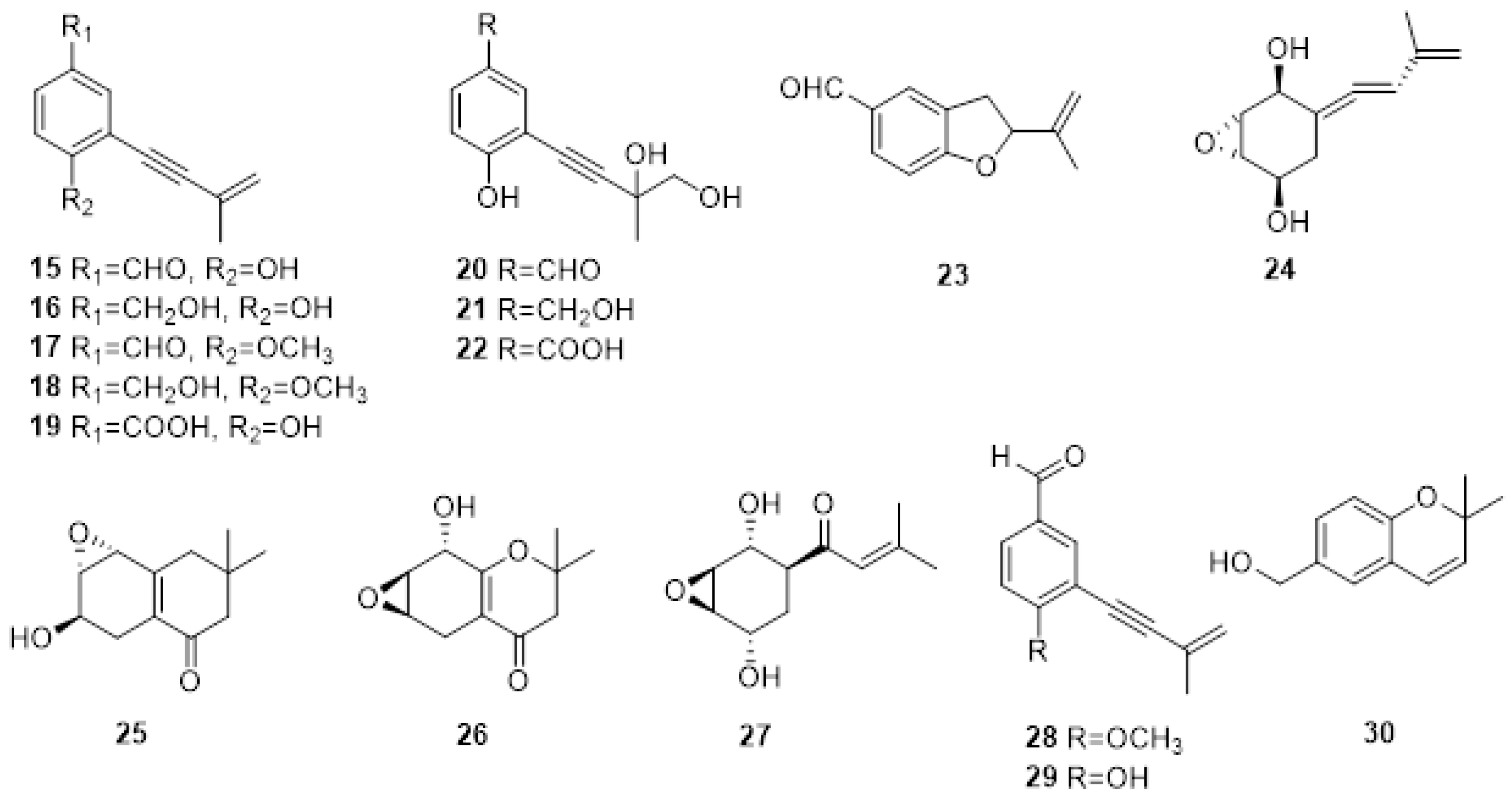
Figure 2. Structure of scytalone eutypine (15), eutypinol (16), O-methyleutypine (17), O-methyleutypinol (18), eutypune carboxylic acid (19), 4-ydroxy-3,4-dihydroxy-3-methylbut-l-ynyl)benzyl alcohol (20), 4-hydroxy-3-(3,4-dihydroxy-3-methylbut-l-ynyl)benzaldehyde (21), 3-hydroxy-3,4-diliydroxy-3-methylbut-I-ynyl)benzoic acid (22), 5-formyl-2-(methylvinyl)[1]benzofuran (23), 5-(3-methylbuta-l.3-dienylidene)-2,3-epoxycyclohexane-l,4-diol (24), 6-hydroxy-2,2-dimethyl-5,6.7,8-tetrahydro-7,8-epoxychroman (25) and 8-hydroxy-2,2-dimethyl-5.6,7,8-tetrahydro-6.7-epoxychroman (26), eutypoxide B (27), eulatinol (28), siccayne (29), eulatachromene (30).
In the same year, the same authors isolated novel allenic epoxycyclohexanes 5-(3-methylbuta-l.3-dienylidene) -2,3-epoxycyclohexane-l,4-diol (24), 6-hydroxy-2,2-dimethyl-5,6.7,8-tetrahydro-7,8-epoxychroman (25) and 8-hydroxy-2,2-dimethyl-5.6,7,8-tetrahydro-6.7-epoxychroman (26) (Figure 2), together with the already reported Eutypine (15). Their structures were established by a combination of spectroscopic, X-ray, and chemical modifications. Again, the most active compound was Eutypine (15). Finally, the authors suggested that compound 24 could be the key intermediate in the transformation of acetylenic compounds into tetrahydrochromanone derivatives [26].
Eutypoxide B (27), biogenetically related to 5-(3-methylbuta-l.3-dienylidene)-2,3-epoxycyclohexane-l,4-diol (24), was isolated from the culture filtrate of E. lata. The structure was confirmed by NMR and X-ray. The authors also conducted a total synthesis of 27, revealing that the key intermediate for the synthesis is a cyclohexanecarbaldehyde compound [28].
Molyneux and co-authors studied three strains of E. lata and their metabolites produced in artificial media by HPLC and GC MS [29]. The novel metabolites eulatinol (28), which is structurally related to the already known siccayne (29), and eulatachromene (30) (Figure 3), a novel chromene analog, were identified by spectroscopic methods as a methoxyquinol derivative. Only one strain produced eutypine (15) in a low amount, whereas the primary metabolite was the corresponding alcohol, eutypinol (16). In the bioassay on grapevine leaf discs, neither eutypinol (15) nor siccayne (29) showed phytotoxicity, whereas eulatinol (28) and eulatachromene (30) caused necrotic spots [29].
Several attempts have been made to identify eutypine from different tissues showing ED symptoms [30][31][32]. In vitro cultures of E. lata isolates produced various SMs, of which eutypine was the main metabolite. However, HPLC analysis of extracts from wood, shoots, and leaves exhibiting symptoms of dieback failed to show the presence of any metabolites [31]. Micropropagated grapevine plantlets treated with crude or purified culture filtrates from nine isolates of E. lata grown on malt yeast broth resulted in various SMs being identified. However, no single compound was consistently detected [32]. A derivative of eutypine, eutypinol, was detected in micropropagated grapevine plantlets inoculated with E. lata mycelium, but PMs were not detected in the sap of vines that had been artificially inoculated with the pathogen [32]. The reasons for the unsuccessful detection of eutypine remain unclear. However, it is possible that following their production; such metabolites are rapidly broken down into compounds that cannot be detected by HPLC [31]. Alternatively, the translocation of metabolites does not occur because the compounds are sufficiently reactive damaging plant tissues in the proximity of fungal infection, reacting in such a way that they are bound irreversibly. Finally, the authors of these studies suggested that as phenolic compounds, the metabolites would be susceptible to oxidative polymerization by plant phenol oxidases, possibly accounting for the dark, wedge-shaped areas typical of E. lata infection [30][31][32].
3. Phytotoxic Metabolites Produced by Fungi Involved in Botryosphaeria Dieback
Botryosphaeriaceae have a cosmopolitan distribution and occur on a wide range of annual and perennial hosts, including grapevines [33][34][35]. They have been described as endophytes or latent pathogens causing serious diseases [35][36]. However, the status of Botryosphaeriaceae species as endophytes in grapevines remains unclear [37]. Botryosphaeriaceae species occur in grape-growing regions of Africa, Asia, Australia, Central America, Europe, and South America [38][39][40][41]. Different species of Botryosphaeriaceae belonging to the genera Botryosphaeria, Diplodia, Dothiorella, Lasiodiplodia, Neofusicoccum, Neoscytalidium, Phaeobotryosphaeria and Spencermartinsia have been reported to be associated with BD of grapevines worldwide. The isolation of S. westrale, S. plurivora, Do. neclivorem, Do. vineagemmae and Do. vidmadera from perennial cankers has brought the total number of Botryosphaeriaceous species isolated from grapevines worldwide to 40 [42][43][44][45][46]. The species infecting grapevines can be classified according to their virulence and can be divided into three different groups, including the highly virulent species, Lasiodiplodia spp. and Neofusicoccum spp., intermediately virulent B. dothidea and Diplodia spp., while Dothiorella spp. and S. viticola are weakly virulent [37].
Low genetic variability is reported from many geographical locations for Botryosphaeria, Diplodia, and Lasiodiplodia species [47]. The first draft of the N. parvum genome was published in 2013, while the first draft genome sequence of D. seriata isolate F98.1 was obtained by Siegwald and co-authors [48]. Studies focused on genetic diversity, the evolution of possible virulence factors, and the pathogenicity of Botryosphaeriaceae species have demonstrated variable levels of virulence between species and isolates [49][50][51][52][53]. All these pathogens have gene clusters involved in the production of carbohydrate-active enzymes (CAZymes), peroxidases, cytochrome P450s, cellular transporters, and secondary metabolism, with a particular focus on toxin production [10][24].
Studies on the phytotoxicity of PMs produced by Botryosphaeriaceae spp. involved in GTDs were first reported by Martos, Andolfi et al. [54]. Species isolated from declining grapevines in Spain (B. dothidea, D. seriata, Do. viticola, N. luteum and N. parvum) produced hydrophilic high-molecular-weight compounds, exopolysaccharides (EPSs) with phytotoxic properties in liquid culture. In addition, lipophilic low molecular weight phytotoxins were isolated from the organic extracts of the culture filtrates of N. luteum, and N. parvum [54].
Among the Botryosphaeriaceae spp. infecting grapevines worldwide, N. parvum and D. seriata are, respectively, the most aggressive and the most widespread worldwide [37], and therefore they are the most studied species producing PMs. Several phenolic dihdroisocoumarins, such as (R)-mellein (31), (3R,4R)-and (3R,4S)-4-hydroxy melleins (32,33) and 5-hydroxymellein (34) (Figure 3) were identified from the organic extract of the culture filtrate of D. seriata [55]. Furthermore, an unknown mellein characterized by NMR was identified as (3R,4R)-4,7- dihydro9mellein (35, Figure 3).

Figure 3 Structure of: (R)-mellein (31), (3R,4R)-and (3R,4S)-4-hydroxy melleins (32, 33), 5-hydroxymellein (34), (3R,4R)-4,7- dihydroxy mellein (35), (+)-terremutin (36), (+)-terremutin hydrate (37), (+)-epi-sphaeropsidone (38), (-)-4-chloro-terremutin hydrate (39), (+)-4—hydroxysuccinate-terremutin hydrate (40), (6R,7R)-asperlin (41), (6R,7S)-dia-asperlin (42), (R)-3-hydroxymellein(43), 6-methyl-salicylic acid (44) and 2-hydroxypropyl salicylic acid (45).
For N. parvum, four phytotoxic metabolites were isolated from organic extracts and identified by spectroscopic and physical examination as isosclerone (2), tyrosol (10, Figure 2), and the previously reported 4-hydroxy-mellein cis and trans mellein (32, 33 Figure 4) [56]. Liquid chromatography-diode array screening of the organic extract of the cultures of 13 isolates of N. parvum resulted in 13 compounds belonging to four different chemical families being identified through spectroscopic analyses and by comparison to previously published literature as detailed in Figure 3: (R)-mellein (31), (3R,4R)-and (3R,4S)-4-hydroxy melleins (32, 33), (+)-terremutin (36), (+)-terremutin hydrate (37) (+)-epi-sphaeropsidone (38), (-)-4-chloro-terremutin hydrate (39), (+)-4—hydroxysuccinate-terremutin hydrate (40), (6R,7R)-asperlin (41), (6R,7S)-dia-asperlin (42), (R)-3-hydroxymellein (43), 6-methyl-salicylic acid (44), 2-hydroxypropyl salicylic acid (45) [57].
(R)-mellein and its derivatives belong to the class of isocoumarines, a class of natural compounds well known to have a wide range of biological activity [58][59]. Moreover, (R)-mellein is a typical PM produced by Botryosphaeriaceae spp. involved in GTD and is phytotoxic to grapevine leaves at different concentrations [55][56].
Ramirez-Suero et al. showed that (R)-mellein could not explain the toxicity of the extracellular organic extract of D. seriata and N. parvum [60]. Purified (R)-mellein was added to the culture medium of calli, but only delayed necrosis and a lower-level expression of defense genes was observed. In addition, the extracellular compounds from N. parvum appeared to be more toxic than those produced by D. seriata. Finally, the authors suggested that it is possible that the pathogenicity of these two fungi depends on synergistic action between the secretion of other types of PMs, such as derivatives of mellein or high molecular weight phytotoxins such as polypeptides or EPSs [60].
The extracellular EPSs produced by an isolate of N. parvum isolated from infected grapevine wood in a vineyard in Spain were biologically and chemically characterized by Cimmino et al. [61]. The EPS was characterized as a mannan having a backbone consisting of (1→6)-linked mannopyranose units, almost all branched at the 2nd position, whereby the arms were composed of 2- and/or 3- linked units. The phytotoxic activity was observed when assayed on grapevine leaves. However, the three replicates of each tested concentration developed symptoms at different times, and differences in the type of symptoms induced were observed; therefore, a conclusion could not be drawn [61].
More recently, purified secreted proteins by N. parvum and D. seriata were assayed on suspension cells of two different Vitis genotypes (V. rupestris and V. vinifera cv. Gewurztraminer) with putative varying susceptibility to BD [62]. The Vitis cells were able to detect secreted proteins produced by Botryosphaeriaceae and respond by producing ROS and by the production of reactive oxygen species and prompt alkalinization of the extracellular medium. Vitis rupestris is characterized by higher medium alkalinization, cell death, and more intense induction of pathogenesis-related genes, whereas V. vinifera cv. Gewurztraminer produced a higher amount of antifungal compound δ-viniferin. The results further suggested that even if the grapevine can react rapidly to BD pathogens, the defense responses are most likely not strong enough to restrict the growth of the pathogen. However, further studies are required to determine the sequences of the secreted proteins and their mode of action [62].
Neofusicoccum australe involved in grapevine decline in Sardinia produced a new cyclohexenone oxide, namely, cyclobotryoxide (46, Figure 4), that was isolated together with 3-methylcatechol (47, Figure 3) and tyrosol. Cyclobotryoxide was the most active metabolite in the different bioassays performed [63].
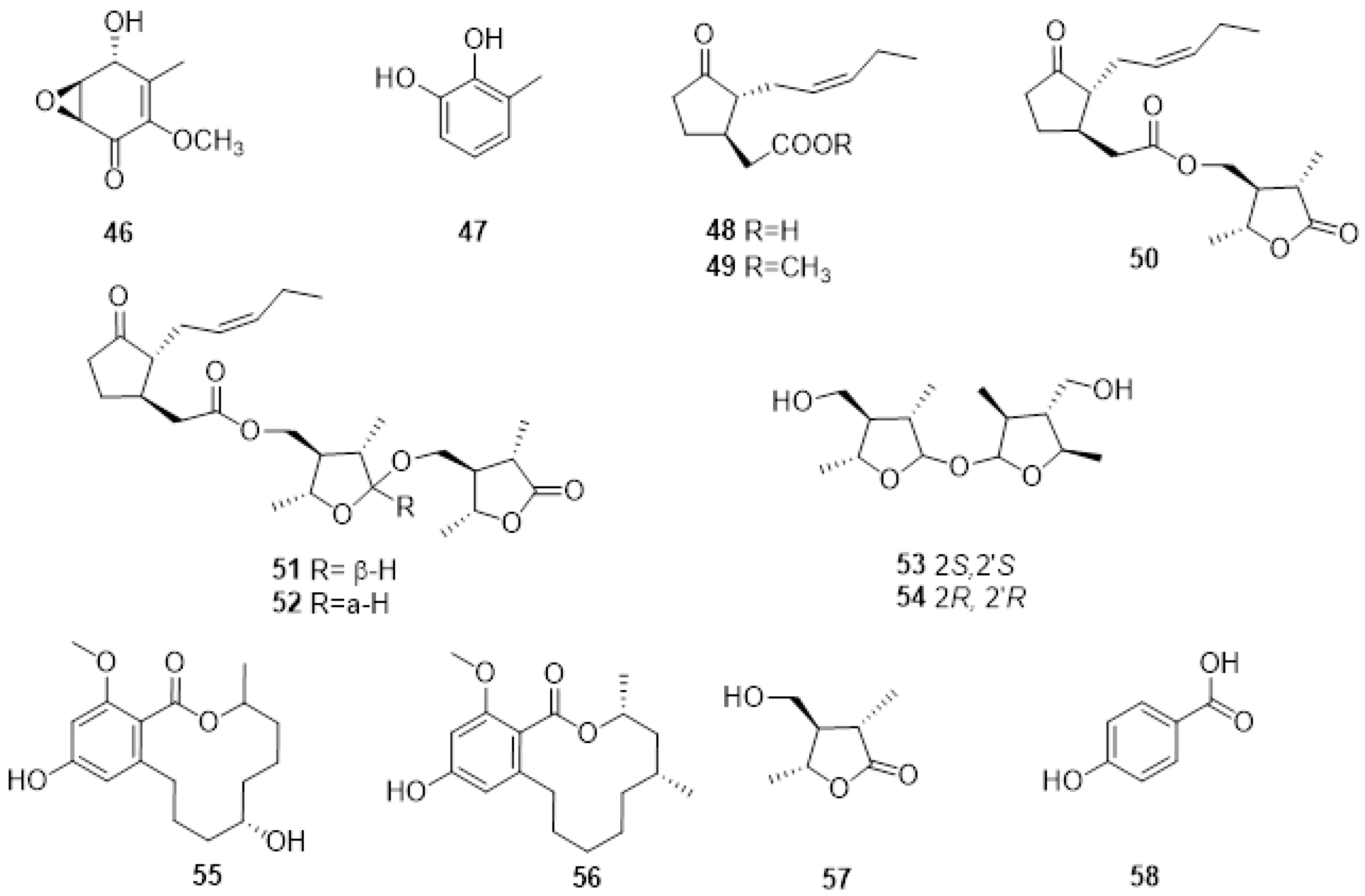
Figure 4. Structure of cyclobotryoxide (46), 3-methylcatechol (47), jasmonic acid (48), jasmonic acid methyl ester (49), lasiojasmonate A (50), lasiojasmonates B and C (51, 52), lasiolactol A and B (53, 54), botryosphaeriodiplodin (55), (5R)-5-hydroxylasiodiplodin (56), (3S,4R,5R)-4-hydroxymethyl-3,5-dimethyldihydro-2-furanone (57) and p-hydroxybenzoic acid (58).
Several endophytic and pathogenic fungi can produce PMs that are also biosynthesized by their host plants [64]. The GTD pathogens, Lasiodiplodia spp., are capable of producing jasmonic acid (48, Figure 4), a known plant hormone, and some of its derivatives when grown in in vitro conditions. Jasmonic acid, its methyl ester (49, Figure 4), and Lasiojasmonate A-C (50-52, Figure 4) were isolated from the grapevine pathogen L. mediterannea [65]. The mode of action of jasmonic acid and lasiojasmonate A as fungal phytotoxins were investigated, and the results suggested that the production of a jasmonic acid derivative such as lasiojasmonate A occurs during the late stages of infection to induce plant jasmonic acid responses such as cell death and to facilitate fungal infection [66][67].
More recently, two novel compounds identified as lasiolactol A and B were isolated and characterised (53, 54 Figure 4) from another strain of L. mediterranea isolated from grapevines in Sicily. These two novel molecules were isolated together with botryosphaeriodiplodin (55, Figure 4), (5R)-5-hydroxylasiodiplodin (56, Figure 4), and (3S,4R,5R)-4-hydroxymethyl-3,5-dimethyldihydro-2-furanone (57, Figure 4), all previously characterized secondary metabolites [68].
References
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39.
- Fontaine, F.; Gramaje, D.; Armengol, J.; Smart, R.; Nagy, Z.A.; Borgo, M.; Rego, C.; Corio-Costet, M.-F. Grapevine Trunk Diseases. A review; OIV Publications: Paris, France, 2016; Volume 24, ISBN 979-10-91799-60-7.
- Gramaje, D.; Mostert, L.; Groenewald, J.Z.; Crous, P.W. Phaeoacremonium: From esca disease to phaeohyphomycosis. Fungal Biol. 2015, 119, 759–783.
- Raimondo, M.L.; Carlucci, A.; Ciccarone, C.; Saddallah, A.; Francesco, L. Identification and pathogenicity of lignicolous fungi associated with grapevine trunk diseases in southern Italy. Phytopathol. Mediter. 2019, 58, 639–662.
- Aigoun-Mouhous, W.; Mahamedi, A.E.; León, M.; Chaouia, C.; Zitouni, A.; Barankova, K.; Eichmeier, A.; Armengol, J.; Gramaje, D.; Berraf-Tebbal, A. Cadophora sabaouae sp. nov. and Phaeoacremonium species associated with Petri disease on grapevine propagation material and young grapevines in Algeria. Plant Dis. 2021, 105, 3657–3668.
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine trunk diseases: A review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant Dis. 2018, 102, 1189–1217.
- Bertsch, C.; Ramirez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clement, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265.
- Blanco-Ulate, B.; Rolshausen, P.; Cantu, D. Draft genome sequence of the ascomycete Phaeoacremonium aleophilum strain UCR-PA7, a causal agent of the esca disease complex in grapevines. Genome Announc. 2013, 1, e00390–e00413.
- Antonielli, L.; Compant, S.; Strauss, J.; Sessitsch, A.; Berger, H. Draft genome sequence of Phaeomoniella chlamydospora strain RR-HG1, a grapevine trunk disease (Esca)-related member of the Ascomycota. Genome Announc. 2014, 2, e00098–e00114.
- Massonnet, M.; Morales-Cruz, A.; Minio, A.; Figueroa-Balderas, R.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Whole-genome resequencing and pan-transcriptome reconstruction highlight the impact of genomic structural variation on secondary metabolism gene clusters in the grapevine Esca pathogen Phaeoacremonium minimum. bioRxiv 2018, 9, 252221.
- Yacoub, A.; Magnin, N.; Gerbore, J.; Haidar, R.; Bruez, E.; Compant, S.; Guyoneaud, R.; Rey, P. The biocontrol root-oomycete, Pythium oligandrum, triggers grapevine resistance and shifts in the transcriptome of the trunk pathogenic fungus, Phaeomoniella chlamydospora. Int. J. Mol. Sci. 2020, 21, 6876.
- Evidente, A.; Bruno, G.; Andolfi, A.; Sparapano, L. Two Naphthalenone Pentakides from Liquid Cultures of Phaeoacremonium aleophilum, a Fungus Associated with Esca of Grapevine. Phytopathol. Mediterr. 2000, 39, 1000–1007.
- Tabacchi, R.; Fkyerat, A.; Poliart, C.; Dubin, G. Phytotoxins from fungi of esca grapevine . Phytopathol. Mediterr. 2000, 1000–1006.
- Abou-Mansour, A.; Tabacchi, R.; Couché, E. Do fungal naphthalenones have a role in the development of esca symptoms? Phytopathol. Mediterr. 2004, 1000–1008.
- Andolfi, A.; Mugnai, L.; Luque, J.; Surico, G.; Cimmino, A.; Evidente, A. Phytotoxins produced by fungi associated with grapevine trunk diseases. Toxins 2011, 3, 1569–1605.
- Medentsev, A.; Akimenko, V. Naphthoquinone metabolites of the fungi. Phytochemistry 1998, 47, 935–959.
- De Gara, L.; de Pinto, M.C.; Tommasi, F. The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol. Biochem. 2003, 41, 863–870.
- Trouillas, F.P.; Urbez-Torres, J.R.; Gubler, W.D. Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 2010, 102, 319–336.
- Trouillas, F.P.; Pitt, W.M.; Sosnowski, M.R.; Huang, R.; Peduto, F.; Loschiavo, A.; Savocchia, S.; Scott, E.S.; Gubler, W.D. Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Div. 2011, 49, 203–223.
- Pitt, W.M.; Trouillas, F.P.; Gubler, W.D.; Savocchia, S.; Sosnowski, M.R. Pathogenicity of diatrypaceous fungi on grapevines in Australia. Plant Dis. 2013, 97, 749–756.
- Sosnowski, M.; Shtienberg, D.; Creaser, M.; Wicks, T.; Lardner, R.; Scott, E. The influence of climate on foliar symptoms of eutypa dieback in grapevines. Phytopathology 2007, 97, 1284–1289.
- Bruez, E.; Baumgartner, K.; Bastien, S.; Travadon, R.; Guérin-Dubrana, L.; Rey, P. Various fungal communities colonise the functional wood tissues of old grapevines externally free from grapevine trunk disease symptoms. Aus. J. Grape Wine Res. 2016, 22, 288–295.
- Blanco-Ulate, B.; Rolshausen, P.E.; Cantu, D. Draft genome sequence of the grapevine dieback fungus Eutypa lata UCR-EL1. Genome Announc. 2013, 1, e00228–e00313.
- Morales-Cruz, A.; Amrine, K.C.; Blanco-Ulate, B.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC genom. 2015, 16, 1–22.
- Onetto, C.A.; Sosnowski, M.R.; Van Den Heuvel, S.; Borneman, A.R. Population genomics of the grapevine pathogen Eutypa lata reveals evidence for population expansion and intraspecific differences in secondary metabolite gene clusters. PLoS Genet. 2022, 18, e1010153.
- Renaud, J.M.; Tsoupras, G.; Stoeckli-Evans, H.; Tabacchi, R. A novel allenic epoxycyclohexane and related compounds from Eutypa lata (Pers: F.) Tul. Helv. Chim. Acta 1989, 72, 1262–1267.
- Renaud, J.M.; Tsoupras, G.; Tabacchi, R. Biologically active natural acetylenic compounds from Eutypa lata (Pers: F.) TUL. Helv. Chim. Acta 1989, 72, 929–932.
- Defrancq, E.; Gordon, J.; Brodard, A.; Tabacchi, R. The synthesis of a novel epoxycyclohexane from the fungus Eutypa lata (Pers: F.) TUL. Helv. Chim. Acta 1992, 75, 276–281.
- Molyneux, R.J.; Mahoney, N.; Bayman, P.; Wong, R.Y.; Meyer, K.; Irelan, N. Eutypa dieback in grapevines: Differential production of acetylenic phenol metabolites by strains of Eutypa lata. J. Agr. Food Chemi. 2002, 50, 1393–1399.
- Rolshausen, P.; Greve, L.; Labavitch, J.; Mahoney, N.; Molyneux, R.; Gubler, W. Pathogenesis of Eutypa lata in grapevine: Identification of virulence factors and biochemical characterization of cordon dieback. Phytopathology 2008, 98, 222–229.
- Mahoney, N.; Molyneux, R.J.; Smith, L.R.; Schoch, T.K.; Rolshausen, P.E.; Gubler, W.D. Dying-arm disease in grapevines: Diagnosis of infection with Eutypa lata by metabolite analysis. J. Agr. Food Chem. 2005, 53, 8148–8155.
- Lardner, R.; Mahoney, N.; Zanker, T.P.; Molyneux, R.J.; Scott, E.S. Secondary metabolite production by the fungal pathogen Eutypa lata: Analysis of extracts from grapevine cultures and detection of those metabolites in planta. Aus. J. Grape Wine Res. 2006, 12, 107.
- Crous, P.W.; Slippers, B.; Wingfield, M.J.; Rheeder, J.; Marasas, W.F.; Philips, A.J.; Alves, A.; Burgess, T.; Barber, P.; Groenewald, J.Z. Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 2006, 55, 235–253.
- Hyde, K.D.; Jones, E.G.; Liu, J.-K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.-Q. Families of dothideomycetes. Fungal Div. 2013, 63, 1–313.
- Slippers, B.; Crous, P.W.; Denman, S.; Coutinho, T.A.; Wingfield, B.D.; Wingfield, M.J. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 2004, 96, 83–101.
- Sakalidis, M.L.; Hardy, G.E.S.; Burgess, T.I. Class III endophytes, clandestine movement amongst hosts and habitats and their potential for disease; a focus on Neofusicoccum australe. Australas. Plant Pathol. 2011, 40, 510–521.
- Urbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, 5–45.
- Urbez-Torres, J.R.; Peduto, F.; Striegler, R.; Urrea-Romero, K.; Rupe, J.; Cartwright, R.; Gubler, W. Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Div. 2012, 52, 169–189.
- Úrbez-Torres, J.; Leavitt, G.; Voegel, T.; Gubler, W. Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Dis. 2006, 90, 1490–1503.
- Pitt, W.M.; Huang, R.; Steel, C.; Savocchia, S. Identification, distribution and current taxonomy of Botryosphaeriaceae species associated with grapevine decline in New South Wales and South Australia. Aus. J. Grape Wine Res. 2010, 16, 258–271.
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Serra, S.; Berraf-Tebbal, A.; Boutiti, M.Z.; Jamâa, M.B.; Phillips, A. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Div. 2015, 71, 201–214.
- Billones-Baaijens, R.; Savocchia, S. A review of Botryosphaeriaceae species associated with grapevine trunk diseases in Australia and New Zealand. Australas. Plant Pathol. 2018, 48, 3–18.
- Pitt, W.M.; Urbez-Torres, J.R.; Trouillas, F.P. Dothiorella and Spencermartinsia, new species and records from grapevines in Australia. Australas. Plant Pathol. 2015, 44, 43–56.
- Comont, G.; Mayet, V.; Corio-Costet, M. First report of Lasiodiplodia viticola, Spencermartinsia viticola and Diplodia intermedia associated with Vitis vinifera grapevine decline in French vineyards. Plant Dis. 2016, 100, 2328.
- Correia, K.C.; Silva, M.A.; de Morais Jr, M.A.; Armengol, J.; Phillips, A.J.; Cômara, M.; Michereff, S.J. Phylogeny, distribution and pathogenicity of Lasiodiplodia species associated with dieback of table grape in the main Brazilian exporting region. Plant Pathol. 2016, 65, 92–103.
- Correia, K.; Silva, M.; Netto, M.; Vieira, W.; Câmara, M.; Michereff, S. First report of grapevine dieback caused by Neoscytalidium hyalinum in Brazil. Plant Dis. 2016, 100, 213.
- Chethana, K.T.; Li, X.; Zhang, W.; Hyde, K.D.; Yan, J. Trail of decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol. Mediterr. 2016, 55, 147.
- Robert-Siegwald, G.; Vallet, J.; Abou-Mansour, E.; Xu, J.; Rey, P.; Bertsch, C.; Rego, C.; Larignon, P.; Fontaine, F.; Lebrun, M.-H. Draft Genome Sequence of Diplodia seriata F98. 1, a Fungal Species Involved in Grapevine Trunk Diseases. Genome Announc. 2017, 5, e00061–e00117.
- Úrbez-Torres, J.; Gubler, W. Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California. Plant Dis. 2009, 93, 584–592.
- Pitt, W.M.; Huang, R.; Steel, C.; Savocchia, S. Pathogenicity and epidemiology of Botryosphaeriaceae species isolated from grapevines in Australia. Australas. Plant Pathol. 2013, 42, 573–582.
- Baskarathevan, J.; Jaspers, M.V.; Jones, E.E.; Cruickshank, R.H.; Ridgway, H.J. Genetic and pathogenic diversity of Neofusicoccum parvum in New Zealand vineyards. Fungal Biol. 2012, 116, 276–288.
- Garcia, J.F.; Lawrence, D.P.; Morales-Cruz, A.; Travadon, R.; Minio, A.; Hernandez-Martinez, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Phylogenomics of plant-associated Botryosphaeriaceae species. Front. Microbiol. 2021, 12, 587.
- Billones-Baaijens, R.; Jones, E.; Ridgway, H.; Jaspers, M. Virulence affected by assay parameters during grapevine pathogenicity studies with Botryosphaeriaceae nursery isolates. Plant Pathol. 2013, 62, 1214–1225.
- Martos, S.; Andolfi, A.; Luque, J.; Mugnai, L.; Surico, G.; Evidente, A. Production of phytotoxic metabolites by five species of Botryosphaeriaceae causing decline on grapevines, with special interest in the species Neofusicoccum luteum and N. parvum. Eur. J. Plant Pathol. 2008, 121, 451–461.
- Djoukeng, J.D.; Polli, S.; Larignon, P.; Abou-Mansour, E. Identification of phytotoxins from Botryosphaeria obtusa, a pathogen of black dead arm disease of grapevine. Eur. J. Plant Pathol. 2009, 124, 303–308.
- Evidente, A.; Punzo, B.; Andolfi, A.; Cimmino, A.; Melck, D.; Luque, J. Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathol. Mediterr. 2010, 49, 74–79.
- Abou-Mansour, E.; Débieux, J.-L.; Ramírez-Suero, M.; Bénard-Gellon, M.; Magnin-Robert, M.; Spagnolo, A.; Chong, J.; Farine, S.; Bertsch, C.; L’Haridon, F. Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry 2015, 115, 207–215.
- Reveglia, P.; Masi, M.; Evidente, A. Melleins—Intriguing Natural Compounds. Biomolecules 2020, 10, 772.
- Sun, H.; Ho, C.L.; Ding, F.; Soehano, I.; Liu, X.-W.; Liang, Z.-X. Synthesis of (R)-mellein by a partially reducing iterative polyketide synthase. J. Am. Chem. Soc. 2012, 134, 11924–11927.
- Ramírez-Suero, M.; Bénard-Gellon, M.; Chong, J.; Laloue, H.; Stempien, E.; Abou-Mansour, E.; Fontaine, F.; Larignon, P.; Mazet-Kieffer, F.; Farine, S. Extracellular compounds produced by fungi associated with Botryosphaeria dieback induce differential defence gene expression patterns and necrosis in Vitis vinifera cv. Chardonnay cells. Protoplasma 2014, 251, 1417–1426.
- Cimmino, A.; Cinelli, T.; Evidente, M.; Masi, M.; Mugnai, L.; Silva, M.A.; Michereff, S.J.; Surico, G.; Evidente, A. Phytotoxic fungal exopolysaccharides produced by fungi involved in grapevine trunk diseases. Nat. Prod. Comm. 2016, 11, 1481–1484.
- Stempien, E.; Goddard, M.-L.; Leva, Y.; Bénard-Gellon, M.; Laloue, H.; Farine, S.; Kieffer-Mazet, F.; Tarnus, C.; Bertsch, C.; Chong, J. Secreted proteins produced by fungi associated with Botryosphaeria dieback trigger distinct defense responses in Vitis vinifera and Vitis rupestris cells. Protoplasma 2018, 255, 613–628.
- Andolfi, A.; Maddau, L.; Cimmino, A.; Linaldeddu, B.T.; Franceschini, A.; Serra, S.; Basso, S.; Melck, D.; Evidente, A. Cyclobotryoxide, a phytotoxic metabolite produced by the plurivorous pathogen Neofusicoccum australe. J. Nat. Prod. 2012, 75, 1785–1791.
- Kusari, S.; Pandey, S.P.; Spiteller, M. Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 2013, 91, 81–87.
- Andolfi, A.; Maddau, L.; Cimmino, A.; Linaldeddu, B.T.; Basso, S.; Deidda, A.; Serra, S.; Evidente, A. Lasiojasmonates A–C, three jasmonic acid esters produced by Lasiodiplodia sp., a grapevine pathogen. Phytochemistry 2014, 103, 145–153.
- Chini, A.; Cimmino, A.; Masi, M.; Reveglia, P.; Nocera, P.; Solano, R.; Evidente, A. The fungal phytotoxin Lasiojasmonate A activates the plant jasmonic acid pathway. J. Exp. Bot. 2018, 69, 3095–3102.
- Reveglia, P.; Chini, A.; Mandoli, A.; Masi, M.; Cimmino, A.; Pescitelli, G.; Evidente, A. Synthesis and mode of action studies of N--S-tyrosin and ester seiridin jasmonate. Phytochemistry 2018, 147, 132–139.
- Andolfi, A.; Basso, S.; Giambra, S.; Conigliaro, G.; Lo Piccolo, S.; Alves, A.; Burruano, S. Lasiolactols A and B produced by the grapevine fungal pathogen Lasiodiplodia mediterranea. Chem. Biodivers. 2016, 13, 395–402.
More
Information
Subjects:
Chemistry, Analytical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
993
Revisions:
2 times
(View History)
Update Date:
16 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




