Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Seiji Shibasaki | -- | 1366 | 2023-01-13 05:28:57 | | | |
| 2 | Camila Xu | Meta information modification | 1366 | 2023-01-13 06:18:47 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Shibasaki, S.; Ueda, M. Molecular Display Technology. Encyclopedia. Available online: https://encyclopedia.pub/entry/40146 (accessed on 07 February 2026).
Shibasaki S, Ueda M. Molecular Display Technology. Encyclopedia. Available at: https://encyclopedia.pub/entry/40146. Accessed February 07, 2026.
Shibasaki, Seiji, Mitsuyoshi Ueda. "Molecular Display Technology" Encyclopedia, https://encyclopedia.pub/entry/40146 (accessed February 07, 2026).
Shibasaki, S., & Ueda, M. (2023, January 13). Molecular Display Technology. In Encyclopedia. https://encyclopedia.pub/entry/40146
Shibasaki, Seiji and Mitsuyoshi Ueda. "Molecular Display Technology." Encyclopedia. Web. 13 January, 2023.
Copy Citation
Molecular display technology or cell-surface engineering is a biotechnological method of genetic engineering that is focused on the cell surface.
yeast
genetic engineering
molecular display technology
1. Introduction
In biochemical studies, yeasts are considered to be a representative model of eukaryotic microbes. The budding yeast Saccharomyces cerevisiae contains only 6611 genes (https://www.yeastgenome.org, accessed on 16 December 2022); however, it has been used for investigating gene and protein functions in recent molecular biology studies [1][2][3][4][5].
Moreover, the use of S. cerevisiae as catalysts in various fermentation industries has a longer history than their application in modern bioscience and biotechnology. For instance, yeasts have been employed in the production processes of fermented foods, such as Japanese sake [6][7], beer [8], bread [9][10], and miso [11][12]. Thus, S. cerevisiae has been related to our diet for a long time and gained a generally regarded as safe (GRAS) status [13][14], although a few strains are pathogenic [15]. Since the advent of genetic engineering, S. cerevisiae has been used for producing valuable compounds. Several recombinant proteins have also been developed as pharmaceuticals using yeast cells [16][17]. Given their usefulness and safety, S. cerevisiae can help us achieve sustainable development.
In this century, biotechnology is expected more than ever to solve socioeconomic issues. Although industrialization has made human life convenient, it has also given rise to environmental issues, such as global climate change, food shortage, and pollution. To overcome these problems, the United Nations proposed 17 Sustainable Development Goals (SDGs) for maintaining human health and establishing a sustainable society [18]. Natural resources or products form the basis of and are consumed during the economic activities in developing and developed countries, causing industrial and environmental problems that need to be mitigated. Considering that biotechnology has improved the quality and scale of industrial and agricultural production, it may also provide several opportunities for sustainable development. The latest biotechnology development in manufacturing not only improves production efficiency, but also promotes international trade and cooperation toward mutual development [19]. Among various biotechnologies, microbial biotechnology is expected to achieve SDGs [20]. Microbiology and microbial technology are considered the key to achieving SDGs via the eradication of infectious diseases, provision of clean water, food security, maintenance of terrestrial and marine biodiversity, and utilization of biofuels [21].
2. General Description of Molecular Display Technology
Molecular display technology or cell-surface engineering is a biotechnological method of genetic engineering that is focused on the cell surface. The phage display system proposed by Smith has the longest history among molecular display systems [22]. It was employed in searching for a clone that can bind to a target compound or investigating protein interactions and is still widely used today [23][24]. A phagemid vector encoding a foreign protein to be displayed as a fusion of the coat protein can be introduced into a phage, resulting in a library consisting of 1012 clones. Next, the panning process is conducted to select positive clones that can bind to target molecules from this phage library; this process includes immobilization of the target molecule on a solid phase, incubation of a library and the target molecule, removal of bound phages from solid phages, infection of Escherichia coli with the recovered phage, and amplification of the clones. This sequential panning cycle should be performed several times. In addition, bacterial cells have been developed and were shown to improve the phage display method, which uses complicated panning processes to isolate a clone capable of binding to a target. For example, surface display systems of foreign proteins using Lactococcus lactis [25], Staphylococcus aureus [26], and E. coli [27] as the host cell have been developed. In addition, unlike phage display systems, bacterial display systems were applied not only in the selection of protein clones that can bind to a target compound, but also in the establishment of whole-cell biocatalysts coupled with metabolic reaction in host cells [28].
Yeasts have been employed in molecular display technology for 20 years [29][30][31]. The yeast S. cerevisiae is useful as a host microorganism in genetic manipulation because it can produce and glycosylate foreign proteins derived from other eukaryotes. This yeast species also has the advantage of high-density cultivation in various media at a low cost. In addition, this yeast not only displays proteins derived from other eukaryotes but also different proteins on its cell surface, i.e., “co-display” [32]. As another characteristic of host cells, several auxotrophic markers can be used in the genetic manipulation of yeast cells for producing different recombinant proteins. Furthermore, flow cytometry or high-throughput screening is applicable for selecting target protein-displaying yeast cells [33][34][35]. Thus, molecular display or cell-surface engineering using the yeast cell surface has many important benefits and practical applications. Yeasts capable of displaying foreign proteins, especially S. cerevisiae, are called “arming yeasts” [36][37][38][39][40]. The principle of molecular display using yeasts and its applications to achieve sustainable development are describe in the subsequent sections.
3. Principle of Molecular Display Technology Using Yeasts
Regardless of the type of host cells selected for molecular display, the anchoring protein must be valid and effective. For example, OmpA has been investigated and used as an anchoring protein to display a foreign protein on the cell surface of E. coli [41][42]. It is usually used as a lipoprotein, which is fused to residues 46–159 of the OmpA porin protein family to anchor to the E. coli cell wall envelope. Moreover, the cA domain of AcmA (a major autolysin from L. lactis) [25] and PgsA from Bacillus subtilis [43] have shown efficacy in displaying several foreign proteins on the cell surface of Lactobacillus sp.
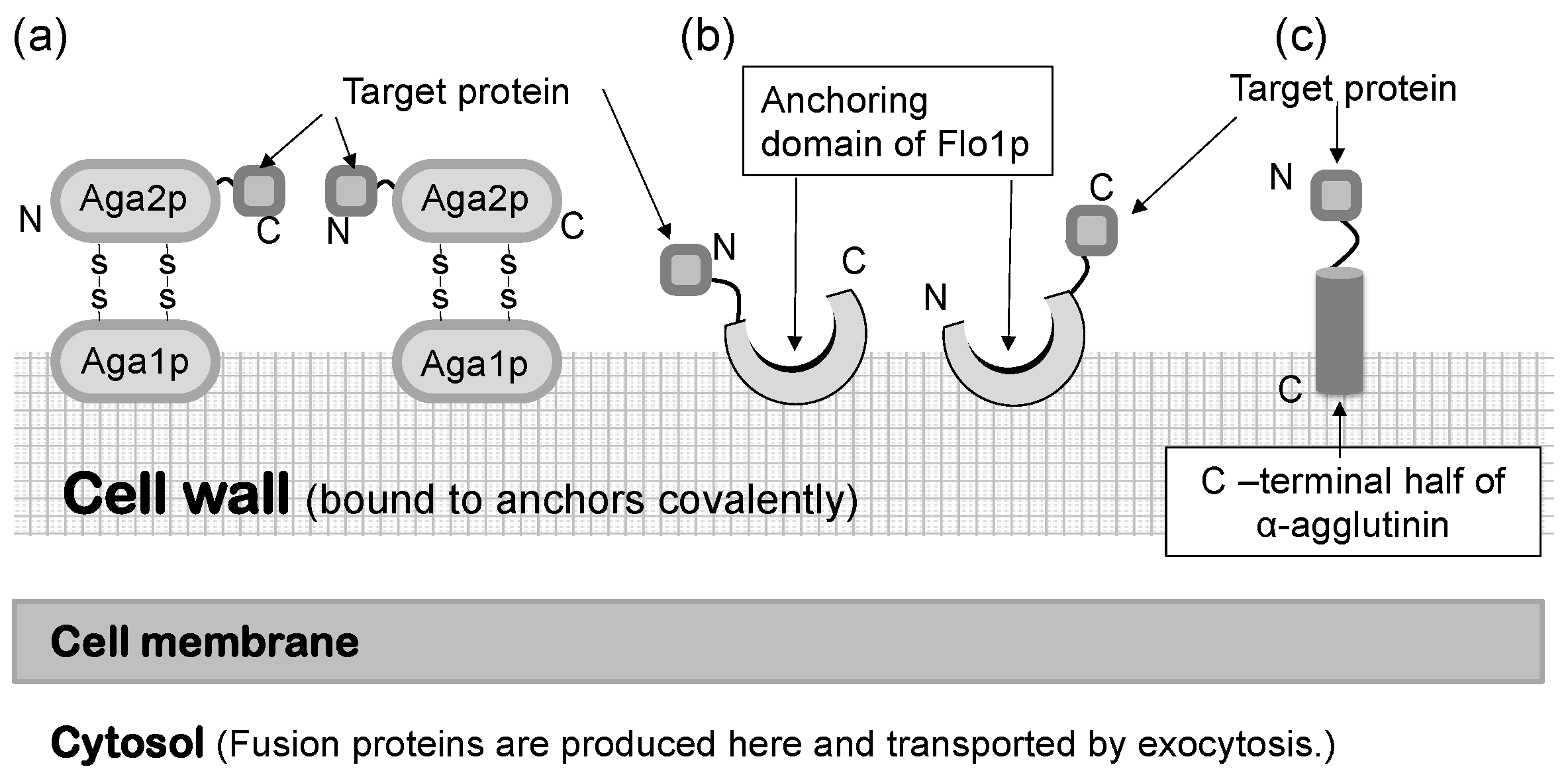
In a S. cerevisiae molecular display system, several anchoring proteins can be used to immobilize the target protein (Figure 1). The Flo1 protein can anchor the target protein in two ways. It is produced and attached on the yeast cell surface for flocculation. In molecular display technology, the target protein to be displayed on the yeast cell surface is usually fused at its N-terminal to the C-terminal of the Flo1 protein. Moreover, Aga1-Aga2 proteins, which are also used for displaying the target protein, are favorable because they can display a C-terminal-free protein on the yeast cell surface. If the active site of the target protein is located in the N-terminal, or if its C-terminal conformation does not affect the function of the target protein, α-agglutinin can be used.
Saccharomyces cerevisiae has a thick, hard cell wall that consists of β-linked glucans and mannoproteins. The cell wall is located outside the plasma membrane and consists of an internal skeletal layer of glucan composed of β-1,3- and β-1,6-linked glucose and a fibrillar or brush-like outer layer composed predominantly of mannoproteins [44]. There are two types of mannoproteins in the thick cell wall [45]. One is loosely bound to the cell wall with non-covalent bonds and can be extracted using sodium dodecylsulfate (SDS). The other mannoprotein can be extracted by using β-1,3- or β-1,6-glucanase. Cell wall proteins illustrated in Figure 1 are glucanase-extractable mannoproteins covalently linked with the glucan layer of the cell wall.
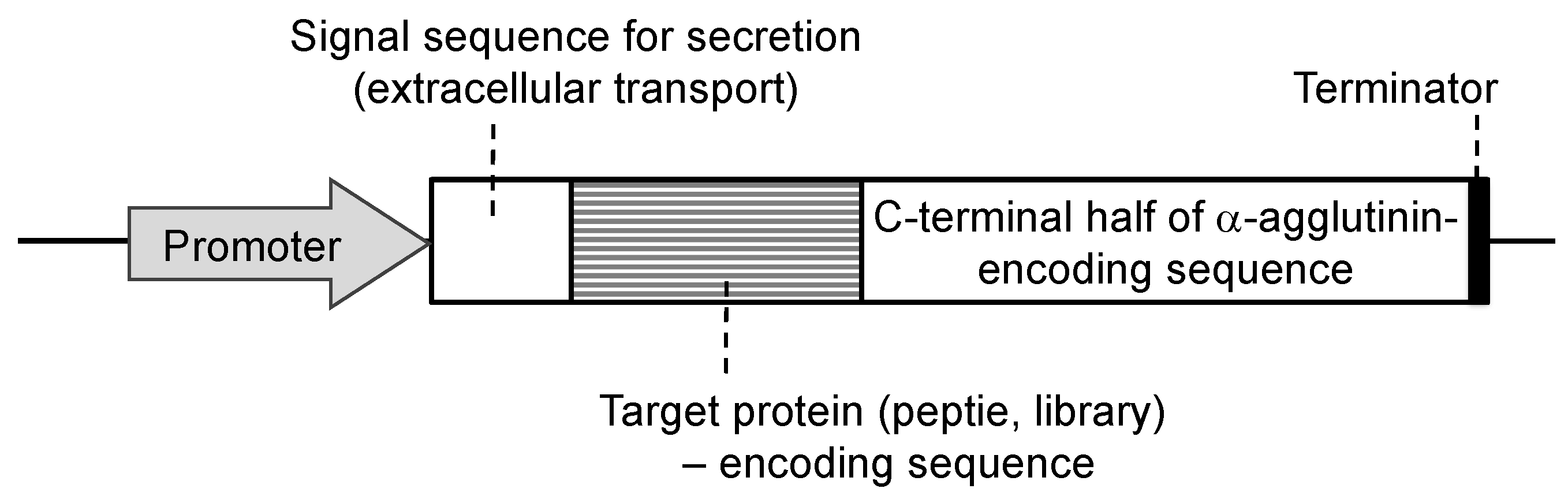
Since the development of yeast molecular display systems, α-agglutinin has been widely investigated and used as an anchoring protein [46][47][48]. Mating-type alpha cells have α-agglutinin protein on their cell surface, which functions during mating with other types of S. cerevisiae cells. This protein is covalently bound to the cell wall, causing the target protein to be fused and stably displayed on the cell surface. Although α-agglutinin itself has a predicted length of 650 amino acids before processing, genetic engineering allows the fusion of the target protein to the C-terminal half (320 amino acids) of α-agglutinin as a cell wall anchor. In the genetic preparation for molecular display, a gene encoding foreign protein is placed between a gene encoding secretion signal sequence and a gene encoding the C-terminal half of α-agglutinin (Figure 2). This genetic construct is expressed under a suitable promoter sequence in a plasmid when it is successfully incorporated by the host yeast cells. In the following sections, several applications of molecular display systems of S. cerevisiae with α-agglutinin are described.

Figure 1. Molecular display systems in Saccharomyces cerevisiae. A target protein is immobilized by several types of anchoring proteins. (a) a-agglutinin-based display system. (b) Flo1p-based display system. (c) α-Agglutinin-based display system. Figures were adapted from [47].

Figure 2. Fusion gene construction for cell-surface display of metal-binding protein and peptide on yeast cell surface. Secretion signal sequence is necessary for extracellular transport of a fusion protein.
References
- Baryshnikova, A.; Costanzo, M.; Dixon, S.; Vizeacoumar, F.J.; Myers, C.L.; Andrews, B.; Boone, C. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010, 470, 145–179.
- Nielsen, J. Yeast systems biology: Model organism and cell factory. Biotechnol. J. 2019, 14, e1800421.
- Coronas-Serna, J.M.; Valenti, M.; Del Val, E.; Fernández-Acero, T.; Rodríguez-Escudero, I.; Mingo, J.; Luna, S.; Torices, L.; Pulido, R.; Molina, M.; et al. Modeling human disease in yeast: Recreating the PI3K-PTEN-Akt signaling pathway in Saccharomyces cerevisiae. Int. Microbiol. 2020, 23, 75–87.
- Vyas, A.; Freitas, A.V.; Ralston, Z.A.; Tang, Z. Fission yeast Schizosaccharomyces pombe: A unicellular “micromammal” model organism. Curr. Protoc. 2021, 1, e151.
- Giga-Hama, Y.; Tohda, H.; Takegawa, K.; Kumagai, H. Schizosaccharomyces pombe minimum genome factory. Biotechnol. Appl. Biochem. 2007, 46, 147–155.
- Kitagaki, H.; Kitamoto, K. Breeding research on sake yeasts in Japan: History recent technological advances and future perspectives. Annu. Rev. Food Sci. Technol. 2013, 4, 215–235.
- Akao, T.; Yashiro, I.; Hosoyama, A.; Kitagaki, H.; Horikawa, H.; Watanabe, D.; Akada, R.; Ando, Y.; Harashima, S.; Inoue, T.; et al. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011, 18, 423–434.
- van Nierop, S.N.; Axcell, B.C.; Cantrell, I.C.; Rautenbach, M. Quality assessment of lager brewery yeast samples and strains using barley malt extracts with anti-yeast activity. Food Microbiol. 2009, 26, 192–196.
- Lahue, C.; Madden, A.A.; Dunn, R.R.; Smukowski Heil, C. History and domestication of Saccharomyces cerevisiae in bread baking. Front. Genet. 2020, 1111, 584718.
- Cobbold, C.A. The rise of alternative bread leavening technologies in the nineteenth century. Ann. Sci. 2018, 75, 21–39.
- Hesseltine, C.W.; Shibasaki, K. Miso. III. Pure culture fermentation with Saccharomyces rouxii. Appl. Microbiol. 1961, 9, 515–518.
- Nakamura, S.; Nakano, Y.; Satoh, H.; Ohtsubo, K. Improved palatability and bio-functionality of super-hard rice by soaking in a barley-koji miso suspension. Biosci. Biotechnol. Biochem. 2013, 77, 2419–2429.
- Sessou, P.; Keisam, S.; Tuikhar, N.; Gagara, M.; Farougou, S.; Jeyaram, K. High-Throughput Illumina MiSeq amplicon sequencing of yeast communities associated with indigenous dairy products from republics of Benin and Niger. Front. Microbiol. 2019, 310, 594.
- van der Hoek, S.A.; Darbani, B.; Zugaj, K.E.; Prabhala, B.K.; Biron, M.B.; Randelovic, M.; Medina, J.B.; Kell, D.B.; Borodina, I. Engineering the yeast Saccharomyces cerevisiae for the production of L-(+)-ergothioneine. Front. Bioeng. Biotechnol. 2019, 117, 262.
- Anoop, V.; Rotaru, S.; Shwed, P.S.; Tayabali, A.F.; Arvanitakis, G. Review of current methods for characterizing virulence and pathogenicity potential of industrial Saccharomyces cerevisiae strains towards humans. FEMS Yeast Res. 2015, 15, fov057.
- Lee, J.; Park, J.S.; Moon, J.Y.; Kim, K.Y.; Moon, H.M. The influence of glycosylation on secretion stability and immunogenicity of recombinant HBV pre-S antigen synthesized in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2003, 303, 427–432.
- Schiller, J.T.; Müller, M. Next generation prophylactic human papillomavirus vaccines. Lancet Oncol. 2015, 16, e217–e225.
- López-Gálvez, F.; Gómez, P.A.; Artés, F.; Artés-Hernández, F.; Aguayo, E. Interactions between microbial food safety and environmental sustainability in the fresh produce supply chain. Foods 2021, 10, 1655.
- Lokko, Y.; Heijde, M.; Schebesta, K.; Scholtès, P.; Van Montagu, M.; Giacca, M. Biotechnology and the bioeconomy–Towards inclusive and sustainable industrial development. New Biotechnol. 2018, 40, 5–10.
- Timmis, K.; de Lorenzo, V.; Verstraete, W.; Ramos, J.L.; Danchin, A.; Brüssow, H.; Singh, B.K.; Timmis, J.K. The contribution of microbial biotechnology to economic growth and employment creation. Microb. Biotechnol. 2017, 10, 1137–1144.
- Fagunwa, O.E.; Olanbiwoninu, A.A. Accelerating the sustainable development goals through microbiology: Some efforts and opportunities. Access Microbiol. 2020, 2, acmi000112.
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317.
- Grimm, S.; Lundberg, E.; Yu, F.; Shibasaki, S.; Vernet, E.; Skogs, M.; Nygren, P.Å.; Gräslund, T. Selection and characterisation of affibody molecules inhibiting the interaction between Ras and Raf in vitro. New Biotechnol. 2010, 27, 766–773.
- von Witting, E.; Lindbo, S.; Lundqvist, M.; Möller, M.; Wisniewski, A.; Kanje, S.; Rockberg, J.; Tegel, H.; Åstrand, M.; Uhlén, M.; et al. Small bispecific affinity proteins for simultaneous target binding and albumin-associated half-life extension. Mol. Pharm. 2021, 18, 328–337.
- Okano, K.; Zhang, Q.; Kimura, S.; Narita, J.; Tanaka, T.; Fukuda, H.; Kondo, A. System using tandem repeats of the cA peptidoglycan-binding domain from Lactococcus lactis for display of both N- and C-terminal fusions on cell surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 2008, 74, 1117–1123.
- Wernérus, H.; Lehtiö, J.; Samuelson, P.; Ståhl, S. Engineering of staphylococcal surfaces for biotechnological applications. J. Biotechnol. 2002, 96, 67–78.
- Salema, V.; Fernández, L.Á. Escherichia coli surface display for the selection of nanobodies. Microb. Biotechnol. 2017, 10, 1468–1484.
- Smith, M.R.; Khera, E.; Wen, F. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 2015, 54, 4021–4032.
- Ueda, M.; Tanaka, A. Genetic immobilization of proteins on the yeast cell surface. Biotechnol. Adv. 2000, 18, 121–140.
- Shibasaki, S.; Maeda, H.; Ueda, M. Molecular display technology using yeast-arming technology. Anal. Sci. 2009, 25, 41–49.
- Shibasaki, S.; Ueda, M. Development of yeast molecular display systems focused on therapeutic proteins, enzymes, and foods: Functional analysis of proteins and its application to bioconversion. Recent Pat. Biotechnol. 2010, 4, 198–213.
- Ito, J.; Kosugi, A.; Tanaka, T.; Kuroda, K.; Shibasaki, S.; Ogino, C.; Ueda, M.; Fukuda, H.; Doi, R.H.; Kondo, A. Regulation of the display ratio of enzymes on the Saccharomyces cerevisiae cell surface by the immunoglobulin G and cellulosomal enzyme binding domains. Appl. Environ. Microbiol. 2009, 75, 4149–4154.
- VanAntwerp, J.J.; Wittrup, K.D. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol. Prog. 2000, 16, 31–37.
- Sun, Y.; Ban, B.; Bradbury, A.; Ansari, G.A.; Blake, D.A. Combining yeast display and competitive FACS to select rare hapten-specific clones from recombinant antibody libraries. Anal. Chem. 2016, 88, 9181–9189.
- Borodina, I.; Jensen, B.M.; Søndergaard, I.; Poulsen, L.K. Display of wasp venom allergens on the cell surface of Saccharomyces cerevisiae. Microb. Cell Fact. 2010, 9, 74.
- Anonymous. Arming yeast with cell-surface catalysts. Chem. Eng. News 1997, 75, 32.
- Ueda, M.; Tanaka, A. Cell surface engineering of yeast: Construction of arming yeast with biocatalyst. J. Biosci. Bioeng. 2000, 90, 125–136.
- Kondo, A.; Ueda, M. Yeast cell-surface display–Applications of molecular display. Appl. Microbiol. Biotechnol. 2004, 64, 28–40.
- Seong, K.T.; Katakura, Y.; Ninomiya, K.; Bito, Y.; Katahira, S.; Kondo, A.; Ueda, M.; Shioya, S. Effect of flocculation on performance of arming yeast in direct ethanol fermentation. Appl. Microbiol. Biotechnol. 2006, 73, 60–66.
- Andreu, C.; Del Olmo, M.L. Yeast arming systems: Pros and cons of different protein anchors and other elements required for display. Appl. Microbiol. Biotechnol. 2018, 102, 2543–2561.
- Nicchi, S.; Giuliani, M.; Giusti, F.; Pancotto, L.; Maione, D.; Delany, I.; Galeotti, C.L.; Brettoni, C. Decorating the surface of Escherichia coli with bacterial lipoproteins: A comparative analysis of different display systems. Microb. Cell Fact. 2021, 20, 33.
- Vahed, M.; Ramezani, F.; Tafakori, V.; Mirbagheri, V.S.; Najafi, A.; Ahmadian, G. Molecular dynamics simulation and experimental study of the surface-display of SPA protein via Lpp-OmpA system for screening of IgG. AMB Express. 2020, 10, 161.
- Narita, J.; Okano, K.; Kitao, T.; Ishida, S.; Sewaki, T.; Sung, M.H.; Fukuda, H.; Kondo, A. Display of alpha-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein and production of lactic acid from starch. Appl. Environ. Microbiol. 2006, 72, 269–275.
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343.
- Cid, V.J.; Durán, A.; del Rey, F.; Snyder, M.P.; Nombela, C.; Sánchez, M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 1995, 59, 345–386.
- Shibasaki, S.; Ueda, M. Bioadsorption strategies with yeast molecular display technology. Biocontrol Sci. 2014, 19, 157–164.
- Ueda, M. Establishment of cell surface engineering and its development. Biosci. Biotechnol. Biochem. 2016, 80, 1243–1253.
- Kipnis, P.; Thomas, N.; Ovalle, R.; Lipke, P.N. The ER-Golgi v-SNARE Bet1p is required for cross-linking alpha-agglutinin to the cell wall in yeast. Microbiology 2004, 150, 3219–3228.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
13 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




