Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Michael N. Romanov | -- | 2535 | 2023-01-05 13:49:04 | | | |
| 2 | Camila Xu | -1 word(s) | 2534 | 2023-01-06 01:41:31 | | | | |
| 3 | Michael N. Romanov | Meta information modification | 2534 | 2023-01-06 08:32:49 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Griffin, D.K.; Larkin, D.M.; O’connor, R.E.; Romanov, M.N. Dinosaurs. Encyclopedia. Available online: https://encyclopedia.pub/entry/39796 (accessed on 28 February 2026).
Griffin DK, Larkin DM, O’connor RE, Romanov MN. Dinosaurs. Encyclopedia. Available at: https://encyclopedia.pub/entry/39796. Accessed February 28, 2026.
Griffin, Darren K., Denis M. Larkin, Rebecca E. O’connor, Michael N. Romanov. "Dinosaurs" Encyclopedia, https://encyclopedia.pub/entry/39796 (accessed February 28, 2026).
Griffin, D.K., Larkin, D.M., O’connor, R.E., & Romanov, M.N. (2023, January 05). Dinosaurs. In Encyclopedia. https://encyclopedia.pub/entry/39796
Griffin, Darren K., et al. "Dinosaurs." Encyclopedia. Web. 05 January, 2023.
Copy Citation
According to Britannica, dinosaurs are described as “Triceratops, contemporary birds, their most recent common ancestor and all of their descendants.” However, for biologists, it could be simpler to picture dinosaurs as reptiles with hind limbs held erect beneath the trunk, similar to how mammals’ hind limbs are held.
dinosaurs
birds
reptiles
chromosome
karyotype
1. Introduction
The question of the origin of reptiles, birds and their relationship to extinct dinosaurs has challenged many generations of biologists; it also continues to interest the lay public. In recent years, this interest has increased due to new paleontological findings and developments in the field of genomics (e.g., [1][2]). In light of recent paleontological findings, the hypothesis that dinosaurs were completely eradicated by the most recent mass extinction event [3][4] has been pervasive in the scientific literature, as well as fiction, film, television, popular culture and the media. However, this scientific dogma has undergone a fundamental revision in recent times; dinosaurs are now thought to be reptile survivors of the most recent extinction event through their evolution into modern birds (e.g., [1][2]). In other words, birds are both reptiles and dinosaurs.
A karyotype represents a map of the genome of interest and every genome sequence assembly would benefit from an accurate cytogenomic map [5]. However, while this can be done directly in extant species by sampling live material, the chromosomal composition of extinct dinosaurs can only be derived by inference. This conclusion can be reached by examining the whole genome chromosome-level assemblies (CLA) of extant species [5]. With information about several species’ CLAs at researchers' disposal, comparative genomics is much more practical in silico [6]. While the auxiliary method of cross-species fluorescence in situ hybridization (zoo-FISH) can uncover further chromosome rearrangements that are difficult to detect using conventional karyotyping (e.g., [7][8][9][10]), comparative genomics enables to outline the genome structure of less well-studied species (e.g., [8][11][12]) and reveal the chromosome rearrangements that led to each species’ distinct karyotype (e.g., [10]) using a reference species as a benchmark, e.g., chickens. The relevance and genomic correlates of such chromosome constituents as evolutionary breakpoint regions (EBRs) and homologous synteny blocks (HSBs) that are features of chromosome evolution [6], as well as the mechanisms behind chromosomal breakage and fusion, can all be addressed with the use of CLAs. Given the prevalence of genomics in modern scientific enquiry, cytogenetics (or, more precisely, cytogenomics) is not only a descriptive discipline but also offers a conceptual framework for the organization of any genome. It also provides an original framework for delineating genome–phenome relationships.
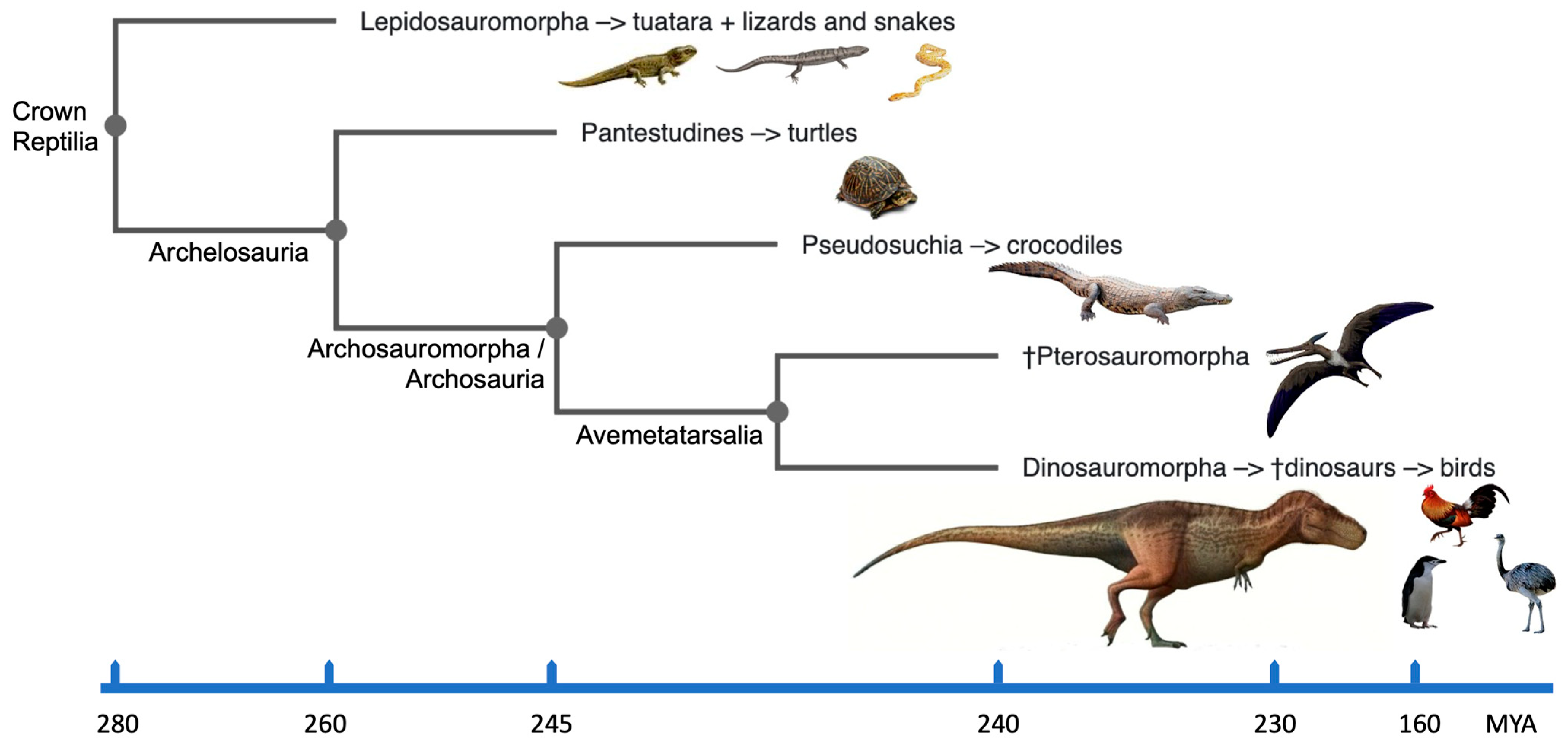
2. Reptilia: Their Phylogeny and Karyotypes
The crown group Reptilia [13] incorporates extinct and existing clades of reptiles, dinosaurs and birds (Figure 1). In particular, it encompasses the diapsid reptiles including the Lepidosauria (tuatara, lizards and snakes) and the Archosauria (extinct dinosaurs, pterosaurs, crocodilians and birds); the latter having originated ~250 million years ago (MYA) [14]. The divergence of synapsids (mammals and their extinct ancestors) in one branch, and anapsids (turtles) and diapsids (other reptiles and birds) in the other, occurred about 310–350 MYA. Evolutionarily, birds represent a monophyletic group of homoeothermic reptiles and are believed to have arisen from theropod dinosaurs about 150 MYA (e.g., [14][15][16]). Archaeopteryx discovered from the late Jurassic (~150 MYA) is recognized as one of the earliest birds. Fossils of most orders of modern birds appear in the early part of the Cenozoic era (65–0 MYA). According to mitochondrial DNA comparisons with extant reptiles, birds are most closely linked to crocodilians, and the divergence between the two lineages is thought to has happened between 210 and 250 MYA (reviewed in [17]). The order Testudines (turtles, tortoises and terrapins) were separated from the Lepidosauria and the Archosauria in the traditional phylogeny because they were thought to be the only survivors of a presumed early anapsid reptile group. The results from molecular phylogeny data estimated from the nucleotide sequences of complete mitochondrial genomes and nuclear genes suggest that turtles should be grouped within the Archelosauria along with crocodilians and birds, while squamates (scaled reptiles including snakes and lizards) are classified into a different clade of Lepidosauria (e.g., [17][18][19][20][21]; Figure 1).

Similar to birds, two main chromosomal components of the karyotypes of snakes, turtles, lizards and tuatara (but not crocodilians) are the macro- and microchromosomes. Snakes have a limited spectrum of karyotypic variation. The diploid number 2n = 36, including 8 pairs of macro- and 10 pairs of microchromosomes, is the most prevalent karyotype in snakes (reviewed in [17]). Lizards also have a low karyotypic variation, mostly with 32–44 chromosomes (e.g., [26][27]) and the extremes being 16 [28] and 62 chromosomes [29]. The diploid number in the lizard Anolis monticola is 48 chromosomes, including 24 macro- and 24 microchromosomes. The fission of chromosomes has been demonstrated in conjunction with lower diploid numbers [30]. The karyotype of the indigenous New Zealand lizard genus Sphenodon (tuatara) has not changed for at least one million years. It has 36 chromosomes, with 14 pairs of macrochromosomes and 4 pairs of microchromosomes. The similarity of the karyotypes of Sphenodon and most Testudines (turtles) points to an ancestral karyotype with a complement of 14 pairs of macrochromosomes and varying numbers of microchromosome pairs [31]. The chromosome number of most crocodilians has long been known [32]; the American alligator (Alligator mississippiensis) karyotype (2n) consists of 32 macrochromosomes, but notably no microchromosomes (in contrast to other reptiles including birds, e.g., [33]). This peculiar feature, unique among reptiles, suggests a derived karyotype arising as a result of wholesale microchromosomal fusion, probably of single origin (given the small number of monophyletic species in which it is observed). Why crocodilians underwent this change and other reptiles did not is unclear.
Using cDNA clones of functional reptile genes and zoo-FISH, Matsuda et al. [17] created comparative cytogenetic maps of the Japanese four-striped rat snake (Elaphe quadrivirgata) and the Chinese soft-shelled turtle (Pelodiscus sinensis). The six biggest chromosomes were found to be near-identical between the chicken and turtle, indicating that chromosome homology was well conserved between the two species. However, compared to the turtle, the snake’s homology to the chicken chromosomes is lower. The chicken Z chromosome shares conserved synteny with the turtle 6q and the snake 2p chromosomes. These findings imply that conserved sequence blocks have survived during the evolution of Testudines and Archosauria in the genomes of turtles and birds. The lineage of snakes has a karyotype with a number of large-sized macrochromosomes and fewer microchromosomes due to a greater frequency of interchromosomal rearrangements that happened between the macrochromosomes and also between macro- and microchromosomes [17]. The suggested that the molecular phylogenetic links between the three genera are supported by the higher conserved synteny in the comparison between the chicken and turtle than in the comparison between the chicken and snake [18][20].
In the 2000s, bacterial artificial chromosome (BAC) libraries became available for the genomes of five reptilian species, American alligator (Alligator mississippiensis), garter snake (Thamnophis sirtalis), tuatara (Sphenodon punctatus), painted turtle (Chrysemys picta) and gila monster (Heloderma suspectum), which represent all five major lineages of extant reptiles [33][34]. The green anole lizard (Anolis carolinensis) was the first reptilian target species for which the genome sequence and CLA were produced [35], with the painted turtle [36], American alligator [37], garter snake [38] and a variety of other reptile species having followed. These advances, along with the progress in avian genomics, make it possible to study the evolutionary relationships and genome history of higher vertebrates (reptiles, birds and mammals) in a broader context [39]. Comparative mapping of birds and reptiles sheds additional light on the amniotes’ evolutionary history [17].
3. Defining Dinosaurs
According to Britannica [40], dinosaurs are described as “Triceratops, contemporary birds, their most recent common ancestor and all of their descendants.” However, for biologists, it could be simpler to picture dinosaurs as reptiles with hind limbs held erect beneath the trunk, similar to how mammals’ hind limbs are held. This sets dinosaurs apart from the majority of other reptiles, including lizards and crocodilians, whose legs are often placed to the side. The related evolutionary clades of dinosaurs, birds and reptiles within the crown group Reptilia [13] are shown in Figure 1. Dinosaurs can therefore be straightforwardly discerned from other animals if its easily identifiable sidelong sister branch of pterosaurs is taken out. With this in mind, dinosaurs are survivors of many extinction events including the most recent Cretaceous–Paleogene (K–Pg) [4]. Data combined from molecular cytogenetics and bioinformatics help demonstrate that their adaptability and capacity to survive extinction events may be due, at least in part, to their karyotypic features.
4. Dinosaurian Forefathers and Avian Heirs
The amniote lineage divided into the reptile/bird lineage (diapsids) and the synapsids, which eventually evolved into mammals (and others), ~325 MYA. Over 17,500 diapsid species exist on the planet, the majority of which are birds (~11,000 species). Turtles (Testudines) diverged first (~255 MYA), followed by crocodilians (~ 252 MYA), pterosaurs (~245 MYA) and then true dinosaurs (including birds) ~240 MYA [41][42]. All of these organisms, including dinosaurs and birds, share a common ancestor (Figure 1) that lived 275 MYA. Dinosaur species remained few in number for the following 30 million years, but during the Jurassic period, their numbers, geographic range and body sizes all increased [43]. The subsequent 135 million years of dinosaur evolution were remarkable because they were the dominant vertebrates on Earth and manifested an extraordinary diversity of species [1]. Amazingly, the dinosaurs survived the catastrophic extinction events of the Carnian–Norian and end-Triassic eras (228 and 201 MYA, respectively). There are currently more than 1000 known species of fossil, with around 30 new species (excluding birds) added each year [44].
Usually, the wide diversity and species abundance of dinosaurs is attributed to the extinction of competing species, which allowed the dinosaurs to prosper. However, it has also been suggested that these remarkable levels of abundance and diversity were a result of dinosaur-specific genetic adaptations, which let them outlive other species in hostile habitats. Examples include unusual bone development rates and highly adapted respiration systems [45], such as unidirectional respiration [46]. Avian species may have evolved successfully due to these types of adaptations; evidence for this may be found in the organization and structure of their genomes.
Multiple bird genome sequencing projects have corrected the important dates of avian diversification, thanks to a revised avian phylogeny based on genome assemblies [47][48]. When the Neognathae (Galloanserae/Neoaves) and the Palaeognathae (Ratites/Tinamous) split apart, this was the time of the first bird evolutionary divergence occurring around 100 MYA. The second divergence occurred when the Galloanserae (Galliformes and Anseriformes) and the Neoaves split 80 MYA, with the divergence of the Galliformes (landfowl, such as chicken, turkey, quail and pheasant) and the Anseriformes (waterfowl, i.e., geese, ducks and swans) occurring around 66 MYA. A further significant split of the Neoaves into the Columbea (including pigeons) and the Passerea (including songbirds) was earlier in evolutionary time (67–69 MYA). Around the time of these two major divergences and after the K–Pg mass extinction event [3][4], a total of 36 neoavian lineages evolved due to diversification in a very brief evolutionary period of 10–15 million years, as shown by Jarvis et al. [48] and Prum et al. [49]. Thus, comparative studies using genomics have revised researchers' understanding of the evolution of dinosaurs, providing fascinating insights into the diversification and the evolution of phenotype [47][48], and prompting further research of the dinosaur karyotype.
5. Characterizing a Hypothetical Dinosaur Genome Organization
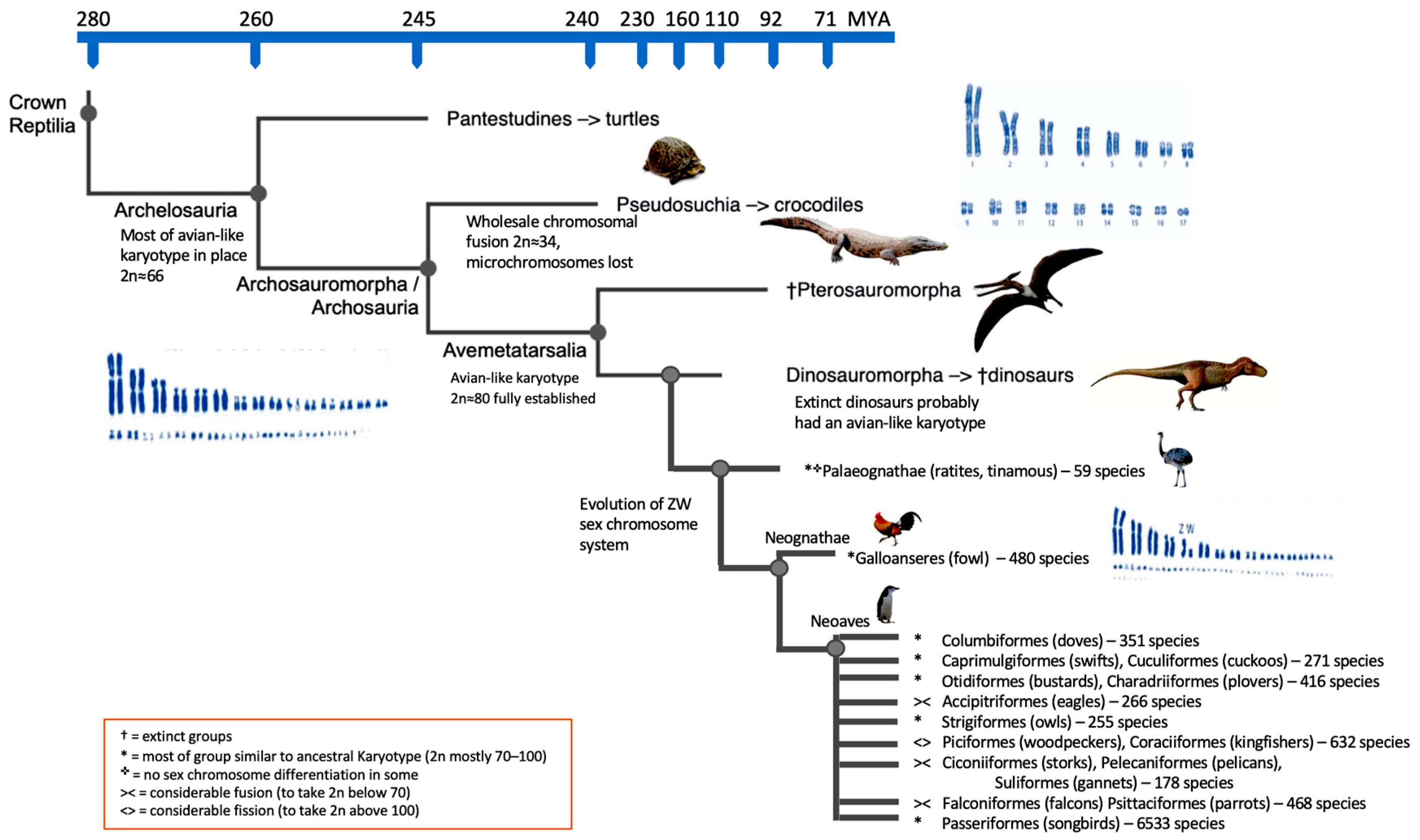
With no intact DNA available from dinosaur fossils, researchers can infer information about extinct dinosaur karyotypes by studying enough avian and reptile CLAs. Romanov et al. [50] were able to determine the most likely ancestral karyotype of all birds by aligning (near) chromosome-level assemblies from six extant birds and an outgroup of the Anolis lizard. This research strategy revealed that the common avian ancestor had a karyotype comparable to that of a chicken or ratite bird [1][50], being a bipedal, terrestrial, tiny Jurassic dinosaur with some flight capacity [1][51]. The next step was to retrace the most likely sequence of rearrangement occurrences that resulted in the avian species’ characteristic karyotypes (e.g., [10]). The zebra finch (Taeniopygia guttata) and budgerigar (Melopsittacus undulatus) were likely subject to the most intra- and interchromosomal changes, while the reconstructed ancestral genome makeup was actually closest to the common chicken karyotype among the birds explored [1][50]. Damas et al. [52] used the method DESCHRAMBLER on fragmented genome assemblies to rebuild the ancestral avian karyotype. A thorough examination of the structure of primitive avian chromosomes was conducted around 14 significant nodes in the evolution of birds. These findings elucidated the varying rates of rearrangement that took place throughout bird evolution. Additionally, it enabled the identification of patterns in the distribution of EBRs along the micro- and macrochromosomes.
A similar method was used by O’Connor et al. [53] to reproduce the diapsids’ most likely ancestral karyotype. A universally hybridizing BAC FISH probe set was created for this purpose [10], which was capable of directly hybridizing across species that diverged hundreds of millions of years ago [54]. The BAC probes used in zoo-FISH investigations produced distinctive signals on the chromosomes of anole lizard (Anolis carolinensis) and further on those of the red-eared slider (Trachemys scripta) and spiny soft-shelled turtle (Apalone spinifera). Based on these zoo-FISH examinations, the chromosome rearrangement events might then be anchored from the viewpoint of an ancestral archelosaur (bird–turtle). The chromosomal modifications from the diapsid ancestor through the archelosaur ancestor [55] and the theropod lineage, and to birds, including chickens, were thus recreated by merging molecular cytogenetics with bioinformatics data [1].
In addition to detecting macro- and microchromosomal homologues, the hybridization of BACs to Trachemys scripta (2n = 50) and Anolis carolinensis (2n = 36) metaphases also revealed the ancestral diapsid karyotype (275 MYA) with 2n = 36–46 and with the ratio of macro- to microchromosomes being approximately 1:1 [1][35][56]. The majority of the key characteristics linked to a typical bird karyotype were already set in the archelosaur progenitor 255 MYA [1][57], which experienced rapid transformation in the preceding 20 million years. This is known because the majority of the Apalone spinifera (2n = 66) and chicken (i.e., ancestral avian) chromosomes (numbered 1-28 + Z) are perfectly syntenic [1]. Studies using chicken chromosome painting on the chromosomes of the painted turtle (Chrysemys picta) [58], red-eared slider (Trachemys scripta; both 2n = 50) [9] and Chinese soft-shelled turtle (Pelodiscus sinensis; 2n = 66) [17] further support the hypothesis that macrochromosomes of birds and turtles are syntenic. Given this information, the only parsimonious explanation is that birds and Pelodiscus sinensis share a common ancestor in terms of their karyotypic structure, as the number of independent convergent events to achieve the same pattern would be statistically extremely unlikely.
To achieve the common avian karyotype pattern from this (~255 MYA) common archelosaur ancestor, to that present in the majority of the main groups of birds, including the Ratites, Galliformes, Anseriformes, Columbea, Passeriformes and others, only about seven fissions would be required. At the rate of chromosomal change occurring at the time, a complete bird-like karyotype would have most likely formed prior to the emergence of the earliest dinosaurs and pterosaurs ~240 MYA. That is, if the same fission rate that had been present for the preceding 20 million years was maintained for another 15 million years, the early dinosaurs probably had bird-like karyotypes [1][59].
The data available therefore strongly imply that not only in most birds, but also with a high degree of certainty, in many, if not most, extinct dinosaurs, the avian chromosomal pattern was maintained mostly unchanged [60]. Figure 2 illustrates this.

Figure 2. Cladogram of the major evolutionary reptilian groups including dinosaurs and several groups of birds. Likely karyotypic changes given, time scale is not linear.
References
- Brusatte, S.L.; Lloyd, G.T.; Wang, S.C.; Norell, M.A. Gradual Assembly of Avian Body Plan Culminated in Rapid Rates of Evolution across the Dinosaur-Bird Transition. Curr. Biol. 2014, 24, 2386–2392.
- Moon, Y.S. The Link between Birds and Dinosaurs: Aves Evolved from Dinosaurs. Korean J. Poult. Sci. 2022, 49, 167–180.
- Schulte, P.; Alegret, L.; Arenillas, I.; Arz, J.A.; Barton, P.J.; Bown, P.R.; Bralower, T.J.; Christeson, G.L.; Claeys, P.; Cockell, C.S.; et al. The Chicxulub Asteroid Impact and Mass Extinction at the Cretaceous-Paleogene Boundary. Science 2010, 327, 1214–1218.
- Berv, J.S.; Field, D.J. Genomic Signature of an Avian Lilliput Effect across the K-Pg Extinction. Syst. Biol. 2018, 67, 1–13.
- Lewin, H.A.; Larkin, D.M.; Pontius, J.; O’Brien, S.J. Every Genome Sequence Needs a Good Map. Genome Res. 2009, 19, 1925–1928.
- Larkin, D.M.; Everts-van der Wind, A.; Rebeiz, M.; Schweitzer, P.A.; Bachman, S.; Green, C.; Wright, C.L.; Campos, E.J.; Benson, L.D.; Edwards, J.; et al. A Cattle–Human Comparative Map Built with Cattle BAC-Ends and Human Genome Sequence. Genome Res. 2003, 13, 1966–1972.
- Nanda, I.; Karl, E.; Griffin, D.K.; Schartl, M.; Schmid, M. Chromosome Repatterning in Three Representative Parrots (Psittaciformes) Inferred from Comparative Chromosome Painting. Cytogenet. Genome Res. 2007, 117, 43–53.
- Modi, W.S.; Romanov, M.; Green, E.D.; Ryder, O. Molecular Cytogenetics of the California Condor: Evolutionary and Conservation Implications. Cytogenet. Genome Res. 2009, 127, 26–32.
- Kasai, F.; O’Brien, P.C.M.; Martin, S.; Ferguson-Smith, M.A. Extensive Homology of Chicken Macrochromosomes in the Karyotypes of Trachemys scripta elegans and Crocodylus niloticus Revealed by Chromosome Painting despite Long Divergence Times. Cytogenet. Genome Res. 2012, 136, 303–307.
- Lithgow, P.E.; O’Connor, R.; Smith, D.; Fonseka, G.; Rathje, C.; Frodsham, R.; O’Brien, P.C.; Ferguson-Smith, M.A.; Skinner, B.M.; Griffin, D.K.; et al. Novel Tools for Characterising Inter- and Intra-chromosomal Rearrangements in Avian Microchromosomes. In Proceedings of the 2014 Meeting on Avian Model Systems, Cold Spring Harbor, NY, USA, 5–8 March 2014; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2014; p. 56.
- Romanov, M.N.; Koriabine, M.; Nefedov, M.; de Jong, P.J.; Ryder, O.A. Construction of a California Condor BAC library and First-generation Chicken–Condor Comparative Physical Map as an Endangered Species Conservation Genomics Resource. Genomics 2006, 88, 711–718.
- Romanov, M.N.; Dodgson, J.B.; Gonser, R.A.; Tuttle, E.M. Comparative BAC-based Mapping in the White-throated Sparrow, a Novel Behavioral Genomics Model, Using Interspecies Overgo Hybridization. BMC Res. Notes 2011, 4, 211.
- Pritchard, A.C.; Nesbitt, S.J. A Bird-like Skull in a Triassic Diapsid Reptile Increases Heterogeneity of the Morphological and Phylogenetic Radiation of Diapsida. R. Soc. Open Sci. 2017, 4, 170499.
- Pereira, S.L.; Baker, A.J. A Mitogenomic Timescale for Birds Detects Variable Phylogenetic Rates of Molecular Evolution and Refutes the Standard Molecular Clock. Mol. Biol. Evol. 2006, 23, 1731–1740.
- Kumar, S.; Hedges, S.B. A Molecular Timescale for Vertebrate Evolution. Nature 1998, 392, 917–920.
- Schmid, M.; Nanda, I.; Hoehn, H.; Schartl, M.; Haaf, T.; Buerstedde, J.M.; Arakawa, H.; Caldwell, R.B.; Weigend, S.; Burt, D.W.; et al. Second Report on Chicken Genes and Chromosomes 2005. Cytogenet. Genome Res. 2005, 109, 415–479.
- Matsuda, Y.; Nishida-Umehara, C.; Tarui, H.; Kuroiwa, A.; Yamada, K.; Isobe, T.; Ando, J.; Fujiwara, A.; Hirao, Y.; Nishimura, O.; et al. Highly conserved linkage homology between birds and turtles: Bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005, 13, 601–615.
- Zardoya, R.; Meyer, A. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc. Natl. Acad. Sci. USA 1998, 95, 14226–14231.
- Hedges, S.B.; Poling, L.L. A Molecular Phylogeny of Reptiles. Science 1999, 283, 998–1001.
- Cao, Y.; Sorenson, M.D.; Kumazawa, Y.; Mindell, D.P.; Hasegawa, M. Phylogenetic Position of Turtles among Amniotes: Evidence from Mitochondrial and Nuclear Genes. Gene 2000, 259, 139–148.
- Cotton, J.A.; Page, R.D.M. Going Nuclear: Gene Family Evolution and Vertebrate Phylogeny Reconciled. Proc. Biol. Sci. 2002, 269, 1555–1561.
- Nesbitt, S.J. The Early Evolution of Archosaurs: Relationships and the Origin of Major Clades. Bull. Am. Mus. Nat. Hist. 2011, 352, 1–292.
- Nesbitt, S.J.; Butler, R.J.; Ezcurra, M.D.; Barrett, P.M.; Stocker, M.R.; Angielczyk, K.D.; Smith, R.M.H.; Sidor, C.A.; Niedźwiedzki, G.; Sennikov, A.G.; et al. The Earliest Bird-line Archosaurs and the Assembly of the Dinosaur Body Plan. Nature 2017, 544, 484–487.
- Lee, M.S.Y. Turtle Origins: Insights from Phylogenetic Retrofitting and Molecular Scaffolds. J. Evol. Biol. 2013, 26, 2729–2738.
- Robinson, O.; Dylus, D.; Dessimoz, C. Phylo.io: Interactive Viewing and Comparison of Large Phylogenetic Trees on the Web. Mol. Biol. Evol. 2016, 33, 2163–2166.
- Lamborot, M. A New Derived and Highly Polymorphic Chromosomal Race of Liolaemus Monticola (Iguanidae) from the ‘Norte Chico’ of Chile. Chromosome Res. 1998, 6, 247–254.
- dos Santos, R.M.; Bertolotto, C.E.; Pellegrino, K.C.; Rodrigues, M.T.; Yonenaga-Yassuda, Y. Chromosomal Studies on Sphaerodactyl Lizards of Genera Gonatodes and Coleodactylus (Squamata, Gekkonidae) Using Differential Staining and Fragile Sites Analyses. Cytogenet. Genome Res. 2003, 103, 128–134.
- Schmid, M.; Feichtinger, W.; Nanda, I.; Schakowski, R.; Visbal Garcia, R.; Manzanilla Puppo, J.; Fernández Badillo, A. An Extraordinarily Low Diploid Chromosome Number in the Reptile Gonatodes taniae (Squamata, Gekkonidae). J. Hered. 1994, 85, 255–260.
- Pellegrino, K.C.; Rodrigues, M.T.; Yonenaga-Yassuda, Y. Chromosomal Polymorphisms Due to Supernumerary Chromosomes and Pericentric Inversions in the Eyelidless Microteiid Lizard Nothobachia ablephara (Squamata, Gymnophthalmidae). Chromosome Res. 1999, 7, 247–254.
- Webster, T.P.; Hall, W.P.; Williams, E.E. Fission in the Evolution of a Lizard Karyotype. Science 1972, 177, 611–613.
- Norris, T.B.; Rickards, G.K.; Daugherty, C.H. Chromosomes of Tuatara, Sphenodon, a Chromosome Heteromorphism and an Archaic Reptilian Karyotype. Cytogenet. Genome Res. 2004, 105, 93–99.
- Cohen, M.M.; Gans, C. The Chromosomes of the Order Crocodilia. Cytogenetics 1970, 9, 81–105.
- Shedlock, A.M.; Botka, C.W.; Zhao, S.; Shetty, J.; Zhang, T.; Liu, J.S.; Deschavanne, P.J.; Edwards, S.V. Phylogenomics of Nonavian Reptiles and the Structure of the Ancestral Amniote Genome. Proc. Natl. Acad. Sci. USA 2007, 104, 2767–2772.
- Wang, Z.; Miyake, T.; Edwards, S.V.; Amemiya, C.T. Tuatara (Sphenodon) Genomics: BAC Library Construction, Sequence Survey, and Application to the DMRT Gene Family. J. Hered. 2006, 97, 541–548.
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The Genome of the Green Anole Lizard and a Comparative Analysis with Birds and Mammals. Nature 2011, 477, 587–591.
- Shaffer, H.B.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K.; et al. The Western Painted Turtle Genome, a Model for the Evolution of Extreme Physiological Adaptations in a Slowly Evolving Lineage. Genome Biol. 2013, 14, R28.
- St John, J.A.; Braun, E.L.; Isberg, S.R.; Miles, L.G.; Chong, A.Y.; Gongora, J.; Dalzell, P.; Moran, C.; Bed’hom, B.; Abzhanov, A.; et al. Sequencing Three Crocodilian Genomes to Illuminate the Evolution of Archosaurs and Amniotes. Genome Biol. 2012, 13, 415.
- Perry, B.W.; Card, D.C.; McGlothlin, J.W.; Pasquesi, G.I.M.; Adams, R.H.; Schield, D.R.; Hales, N.R.; Corbin, A.B.; Demuth, J.P.; Hoffmann, F.G.; et al. Molecular Adaptations for Sensing and Securing Prey and Insight into Amniote Genome Diversity from the Garter Snake Genome. Genome Biol. Evol. 2018, 10, 2110–2129.
- Modi, W.S.; Crews, D. Sex Chromosomes and Sex Determination in Reptiles. Curr. Opin. Genet. Dev. 2005, 15, 660–665.
- Dinosaur Ancestors. Encyclopedia Britannica. Available online: https://www.britannica.com/animal/dinosaur/Dinosaur-ancestors (accessed on 24 October 2022).
- Shedlock, A.M.; Edwards, S.V. Amniotes (Amniota). In The Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: Oxford, UK, 2009; pp. 375–379.
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of Life Reveals Clock-Like Speciation and Diversification. Mol. Biol. Evol. 2015, 32, 835–845.
- Benton, M.J.; Forth, J.; Langer, M.C. Models for the Rise of the Dinosaurs. Curr. Biol. 2014, 24, R87–R95.
- Weishampel, D.O. The Dinosauria, 2nd ed.; University of California Press: Berkeley, CA, USA, 2004.
- O’Connor, P.M.; Claessens, L.P.A.M. Basic Avian Pulmonary Design and Flow-through Ventilation in on-avian Theropod Dinosaurs. Nature 2005, 436, 253–256.
- Farmer, C.G.; Sanders, K. Unidirectional Airflow in the Lungs of Alligators. Science 2010, 327, 338–340.
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative Genomics Reveals Insights into Avian Genome Evolution and Adaptation. Science 2014, 346, 1311–1320.
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome Analyses Resolve Early Branches in the Tree of Life of Modern Birds. Science 2014, 346, 1320–1331.
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A Comprehensive Phylogeny of Birds (Aves) Using Targeted Next-generation DNA Sequencing. Nature 2015, 526, 569–573.
- Romanov, M.N.; Farré, M.; Lithgow, P.E.; Fowler, K.E.; Skinner, B.M.; O’Connor, R.; Fonseka, G.; Backström, N.; Matsuda, Y.; Nishida, C.; et al. Reconstruction of Gross Avian Genome Structure, Organization and Evolution Suggests That the Chicken Lineage Most Closely Resembles the Dinosaur Avian Ancestor. BMC Genom. 2014, 15, 1060.
- Witmer, L.M. The Debate on Avian Ancestry: Phylogeny, Function, and Fossils. In Mesozoic Birds: Above the Heads of Dinosaurs; Chiappe, L.M., Witmer, L.M., Eds.; University of California Press: Berkeley, CA, USA, 2002; pp. 3–30.
- Damas, J.; Kim, J.; Farré, M.; Griffin, D.K.; Larkin, D.M. Reconstruction of Avian Ancestral Karyotypes Reveals Differences in the Evolutionary History of Macro- and Microchromosomes. Genome Biol. 2018, 19, 155.
- O’Connor, R.E.; Romanov, M.N.; Kiazim, L.G.; Barrett, P.M.; Farré, M.; Damas, J.; Ferguson-Smith, M.; Valenzuela, N.; Larkin, D.M.; Griffin, D.K. Reconstruction of the Diapsid Ancestral Genome Permits Chromosome Evolution Tracing in Avian and Non-avian Dinosaurs. Nat. Commun. 2018, 9, 1883.
- Damas, J.; O’Connor, R.; Farré, M.; Lenis, V.P.E.; Martell, H.J.; Mandawala, A.; Fowler, K.; Joseph, S.; Swain, M.T.; Griffin, D.K.; et al. Upgrading Short-read Animal Genome Assemblies to Chromosome Level Using Comparative Genomics and a Universal Probe Set. Genome Res. 2017, 27, 875–884.
- Benton, M.J.; Donoghue, P.C.J.; Asher, R.J.; Friedman, M.; Near, T.J.; Vinther, J. Constraints on the Timescale of Animal Evolutionary History. Palaeontol. Electron. 2015, 18, 1–106.
- Beçak, W.; Beçak, M.L.; Nazareth, H.R.S.; Ohno, S. Close Karyological Kinship Between the Reptilian Suborder Serpentes and the Class Aves. Chromosoma 1964, 15, 606–617.
- Uno, Y.; Nishida, C.; Tarui, H.; Ishishita, S.; Takagi, C.; Nishimura, O.; Ishijima, J.; Ota, H.; Kosaka, A.; Matsubara, K.; et al. Inference of the Protokaryotypes of Amniotes and Tetrapods and the Evolutionary Processes of Microchromosomes from Comparative Gene Mapping. PLoS ONE 2012, 7, e53027.
- Badenhorst, D.; Hillier, L.W.; Literman, R.; Montiel, E.E.; Radhakrishnan, S.; Shen, Y.; Minx, P.; Janes, D.E.; Warren, W.C.; Edwards, S.V.; et al. Physical Mapping and Refinement of the Painted Turtle Genome (Chrysemys picta) Inform Amniote Genome Evolution and Challenge Turtle-Bird Chromosomal Conservation. Genome Biol. Evol. 2015, 7, 2038–2050.
- Baron, M.G.; Norman, D.B.; Barrett, P.M. A New Hypothesis of Dinosaur Relationships and Early Dinosaur Evolution. Nature 2017, 543, 501–506.
- O’Connor, R.E.; Kiazim, L.; Skinner, B.; Fonseka, G.; Joseph, S.; Jennings, R.; Larkin, D.M.; Griffin, D.K. Patterns of Microchromosome Organization Remain Highly Conserved Throughout Avian Evolution. Chromosoma 2019, 128, 21–29.
More
Information
Subjects:
Evolutionary Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
3.8K
Revisions:
3 times
(View History)
Update Date:
06 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





