Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Guendalina Lucarini | -- | 1531 | 2022-12-30 22:03:02 | | | |
| 2 | Beatrix Zheng | Meta information modification | 1531 | 2023-01-03 02:17:11 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rizzetto, G.; Lucarini, G.; Simoni, E.D.; Molinelli, E.; Mattioli-Belmonte, M.; Offidani, A.; Simonetti, O. Tissue Biomarkers Predicting Status in Cutaneous Melanoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/39641 (accessed on 07 February 2026).
Rizzetto G, Lucarini G, Simoni ED, Molinelli E, Mattioli-Belmonte M, Offidani A, et al. Tissue Biomarkers Predicting Status in Cutaneous Melanoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/39641. Accessed February 07, 2026.
Rizzetto, Giulio, Guendalina Lucarini, Edoardo De Simoni, Elisa Molinelli, Monica Mattioli-Belmonte, Annamaria Offidani, Oriana Simonetti. "Tissue Biomarkers Predicting Status in Cutaneous Melanoma" Encyclopedia, https://encyclopedia.pub/entry/39641 (accessed February 07, 2026).
Rizzetto, G., Lucarini, G., Simoni, E.D., Molinelli, E., Mattioli-Belmonte, M., Offidani, A., & Simonetti, O. (2022, December 30). Tissue Biomarkers Predicting Status in Cutaneous Melanoma. In Encyclopedia. https://encyclopedia.pub/entry/39641
Rizzetto, Giulio, et al. "Tissue Biomarkers Predicting Status in Cutaneous Melanoma." Encyclopedia. Web. 30 December, 2022.
Copy Citation
Cutaneous melanoma is a severe neoplasm that shows early invasiveness of the lymph nodes draining the primary site, with increased risk of distant metastases and recurrence. The tissue biomarker identification could be a new frontier to predict the risk of early lymph node invasiveness, especially in cases considered by current guidelines to be at low risk of lymph node involvement and not requiring evaluation of the sentinel lymph node (SLN).
melanoma
lymph nodes
biomarker
1. Lymphovascular Invasion (LVI)
In the literature, it is reported that lymphovascular invasion (LVI), which consists of the finding during routine histological examination of melanoma tumor cells within lymphatic or hematic vessels, correlates with a worse clinical outcome [1]. The use of highly specific and sensitive antibody biomarkers for melanoma (S-100, Mart-1), for cell proliferation (Ki-67, PCNA), and for lymphatic vessels (LYVE-1, D2-40) allowed a better assessment of LVI. All these biomarkers were assessed by immunohistochemistry directly on primary melanoma tissue. In addition, other parameters that are better analyzed with these biomarkers are lymphatic vessel density (LVD) and peritumoral lymphatic new vessel formation [2][3]. Specifically, LVI was estimated to be present in 50% of LVI-negative patients with hematoxylin/eosin staining after immunohistochemistry (IHC) [4][5][6].
Some studies suggest that intratumoral or peritumoral LVI and LVD may correlate with sentinel node loco-regional lymph node metastasis and patient survival, but the results are not unequivocal [4][7][8][9][10].
2. VEGF
Lymphangiogenesis is a necessary condition for tumor involvement of the lymphatic and lymph node system. Glycoproteins of the vascular endothelium growth factor (VEGF) family, produced by tumor cells themselves but also tumor-associated fibroblasts and tumor-associated macrophages, help stimulate lymphangiogenesis [11]. VEGF also acts both on endothelial cells and in an autocrine way on tumor cells by promoting their survival and proliferation [12].
The assessment through immunohistochemistry on melanoma tissue of the biomarkers VEGF-C, VEGF-D and the VEGFR receptor to predict SLN positivity has been proposed in the literature, but the results are currently inconsistent. A study by Gallego et al. [13] showed that VEGF-C in the peritumoral versus intratumoral site is associated with SLN positivity. The use of immunohistochemistry and polymerase chain reaction (PCR) for VEGF-C, VEGF-D, and VEGFR-3 proved to be a good predictor of SLN positivity [10][14]. However, other studies have not shown an association among VEGF-A, VEGF-D, or VEGFR receptors [15][16].
Recently, a study by Toberer et al. [17] showed in a statistically significant manner in a sample of 58 patients that VEGFR-3 expression is an independent predictor of SLN positivity and could be used in the future when choosing to perform SLN. VEGF-A was also statistically significantly shown to be more highly expressed in the tumor tissue of SLN-positive patients; however, on binary logistic regression, it did not prove to be an independent factor for SLN outcome.
The ambiguous results require further multicenter prospective studies to better clarify the role of VEGF and VEGFR in predicting SLN involvement.
3. Tetraspanin CD9
Tetraspanin CD9 is a transmembrane protein that plays a key role in tumor progression, as it functions as a metastasis suppressor in some neoplasms [18]. Specifically, its reduced expression correlates with metastatic progression of the malignancy and poor prognosis. CD9, when overexpressed, enhanced melanocyte motility, suggesting that its overexpression may partly cause the invasion activity of melanoma cells across the Matrigel [19][20].
A recent study [21] conducted on cutaneous melanoma evaluated CD9 expression by immunohistochemistry and immunofluorescence, showing that CD9 was not expressed in thin melanomas, whereas it reappeared in 46% of intermediate and thick melanomas at specific areas of invasion, near or within blood or lymphatic vessels. All of these CD9 stained tumors showed a positive SLN, highlighting that CD9 expression may be a strong predictor of SNB positivity and, therefore, an excellent biomarker for assessing SLN status.
CD9 is also uniformly expressed in melanocytic nevi, leading people to initially consider a protective role of this protein. However, in invasive melanoma, as already seen in other tumors [22][23][24][25][26][27], an increase in CD9 correlates with lymph node metastases, distant metastases, and a worse outcome [21]. This can be explained by considering that tetraspanins can act as both suppressors and promoters of metastases depending on the status of the cell membrane and vesicular structures [28]. Some vitro studies on transendothelial migration of melanoma cells showed a significant role of CD9 in tumor–endothelial/lymphatic cell interaction and vascular dissemination of tumor cells [29]. Therefore, the role of CD9 in determining SLN status seems interesting, as also confirmed by Erovic et al. [30] showing that this tetraspanin is a valid marker for lymphatic endothelial cells and able to promote transmigration of tumor cells through the adherence to lymphatic vessels (Figure 1). Further studies are needed to evaluate its role as a biomarker of the SLN especially in patients with intermediate to thick melanoma.
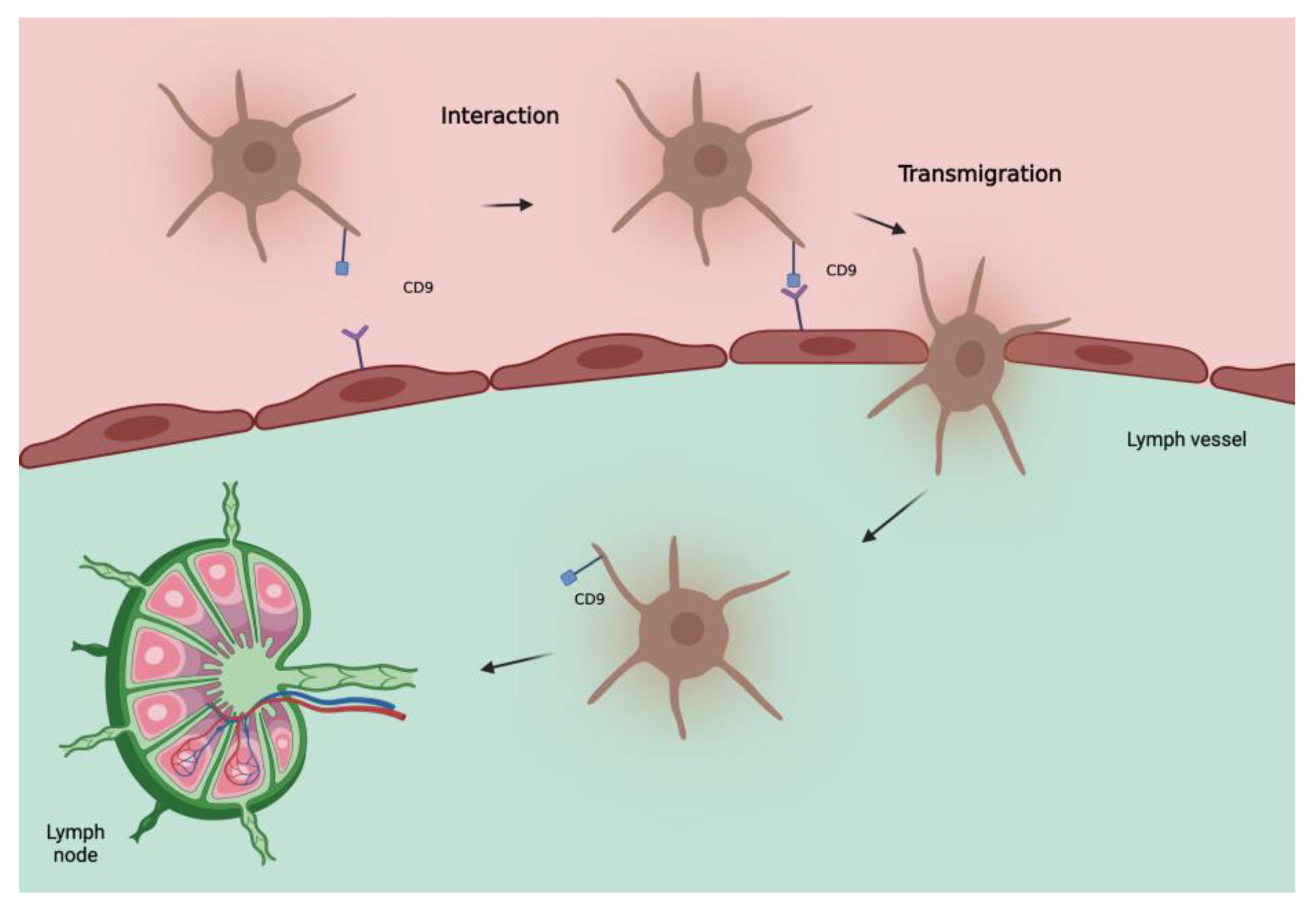
Figure 1. Hypothetical model of involvement of tetraspanin CD9 in metastatic spread of melanoma cells to lymph nodes. CD9, localized at the melanoma cell-endothelial/lymphatic cell contact area during active transmigration of tumor cells across endothelial/lymphatic vessels, facilitates lymph node invasion.
4. LYVE-1 and D2-40
Lymphatic vessel endothelial hyaluronan receptor-1 (LYVE-1), a selective marker of lymphatic vessels, and D2-40, an endothelial marker known as podoplanin, were used to assess LVI in cutaneous melanomas. Their distribution was assessed by immunohistochemistry directly on primary melanoma tissue. Using both of these biomarkers, no correlation was found between LVI and SLN positivity [31]. However, if LVI was associated with the presence of intratumoral lymphatic vessels, the possibility of predicting SLN status increased, with a positive predictive value of 80% and a negative predictive value of 72%.
D2-40 was used alone to assess LVI in cutaneous melanoma <2 mm, showing good predictive ability for positive SLN [32]. These data suggest that LVI assessment implemented using LYVE-1 and D2-40 may be an indicator for SLN status.
5. Gene Expression Profile Test (31-GEP)
31-GEP is a test used to stratify the risk of SLN positivity, identifying high-risk (>5%) patients who are candidates for SLN biopsy. 31-GEP allows evaluation of molecular expression signatures to guide staging in patients with melanoma [33][34][35][36][37][38].
31-GEP is performed on a sample of primary melanoma tissue normally fixed in formalin and embedded in paraffin. Real-time polymerase chain reaction (RT-PCR) is used for the evaluation of expressed genes. From a clinical point of view, this method can be used at the same time as the histological examination. 31-GEP is based on evaluating, directly on the melanoma tissue sample, the expression of gene levels of a panel containing several genes. Specifically, markers can be found for cell migration/chemotaxis/metastasis, secretory molecules, adhesion, lymphocyte invasion, transcription factors, differentiation/proliferation structural proteins, and surface receptors (CXCL14, SPP1, CLCA2, S100A9, S100A8, BAP-1, MGP, GJA1, DSC1, PPL, LTA4H, TRIM29, KRT6B, KRT14, CRABP2, SPRRIB, TACSTD2, CLCA2, ROBO1, CST6, SAP130, ID2, EIF1B, ARG1, AQP1, RBM23, and TYRP1) [39].
Genetic tests can identify a class 1 low risk of SLN positivity or a class 2 high risk. Class 1 is considered to have a very low risk of both metastasis and mortality, also providing greater tranquillity for patients and improving their quality of life [37].
DecisionDx-Melanoma has recently been proposed as an additional risk assessment parameter for SLN positivity, integrating the 31-GEP test with other clinical/pathological aspects for the evaluation of SLN in patients with thin melanoma [40][41].
This could, in the near future, introduce a new variable to be considered, highlighting population groups with a >5% risk of SLN positivity, optimizing the treatment.
All the melanoma tissue biomarkers are summarized in Table 1.
Table 1. summary of reviewed biomarkers for SLN positivity.
| Biomarker | Rationale | Sample | Results |
|---|---|---|---|
| VEGFR-3 Toberer et al. [17] |
Involved in the stimulation of lymphangiogenesis | 58 patients |
|
| Tetraspanin CD9 Lucarini et al. [21] |
Transmembrane protein, key role in tumor progression Both suppressor and promoter of metastases, depending on the status of the cell membrane and vesicular structures |
140 patients Melanocytic nevus 20 Primary melanoma 120
|
|
| LYVE-1 Doeden et al. [31] |
LYVE-1 selective marker of lymphatic vessels | 94 patients |
|
| D2-40 Fohn et al. [32] |
D2-40 endothelial marker (podoplanin) In combination, better histological definition of LVI |
158 patients Primary melanomas ≤ 2.0 mm |
|
| 31-GEP Vetto et al. [39] |
Gene expression profile test, (markers for cell migration/chemotaxis/metastasis, secretory molecules, adhesion, lymphocyte invasion, transcription factors, differentiation/proliferation structural proteins and surface receptors) Identifies high-risk (>5%) patients, candidates for SLN |
690 patients (total validation cohort, retrospective) staged I–III follow-up median 7 years 1421 patients (prospectively-tested) |
|
Vascular endothelium growth factor receptor-3 (VEGF), statistically significant (ss), lymphatic vessel endothelial hyaluronan receptor-1 (LYVE-1), lymphovascular invasion (LVI), sentinel lymph node (SLN), and gene expression profile test (31-GEP).
References
- Scoggins, C.R.; Ross, M.I.; Reintgen, D.S.; Noyes, R.D.; Goydos, J.S.; Beitsch, P.D.; Urist, M.M.; Ariyan, S.; Davidson, B.S.; Sussman, J.J.; et al. Prospective Multi-Institutional Study of Reverse Transcriptase Polymerase Chain Reaction for Molecular Staging of Melanoma. J. Clin. Oncol. 2006, 24, 2849–2857.
- Ji, R.-C. Lymphatic endothelial cells, tumor lymphangiogenesis and metastasis: New insights into intratumoral and peritumoral lymphatics. Cancer Metastasis Rev. 2006, 25, 677–694.
- Xu, X.; Gimotty, P.A.; Guerry, D.; Karakousis, G.; Van Belle, P.; Liang, H.; Montone, K.; Pasha, T.; Ming, M.E.; Acs, G.; et al. Lymphatic invasion revealed by multispectral imaging is common in primary melanomas and associates with prognosis. Hum. Pathol. 2008, 39, 901–909.
- Petersson, F.; Diwan, A.H.; Ivan, D.; Gershenwald, J.E.; Johnson, M.M.; Harrell, R.; Prieto, V.G. Immunohistochemical detection of lymphovascular invasion with D2-40 in melanoma correlates with sentinel lymph node status, metastasis and survival. J. Cutan. Pathol. 2009, 36, 1157–1163.
- Petitt, M.; Allison, A.; Shimoni, T.; Uchida, T.; Raimer, S.; Kelly, B. Lymphatic invasion detected by D2-40/S-100 dual immunohistochemistry does not predict sentinel lymph node status in melanoma. J. Am. Acad. Dermatol. 2009, 61, 819–828.
- Rose, A.E.; Christos, P.J.; Lackaye, D.; Shapiro, R.L.; Berman, R.; Mazumdar, M.; Kamino, H.; Osman, I.; Darvishian, F. Clinical Relevance of Detection of Lymphovascular Invasion in Primary Melanoma Using Endothelial Markers D2-40 and CD34. Am. J. Surg. Pathol. 2011, 35, 1441–1449.
- Dadras, S.S.; Paul, T.; Bertoncini, J.; Brown, L.F.; Muzikansky, A.; Jackson, D.G.; Ellwanger, U.; Garbe, C.; Mihm, M.C.; Detmar, M. Tumor lymphangiogenesis: A novel prognostic indicator for cutaneous melanoma metastasis and survival. Am. J. Pathol. 2003, 162, 1951–1960.
- Huber, G.F.; Fritzsche, F.R.; Züllig, L.; Storz, M.; Graf, N.; Haerle, S.K.; Jochum, W.; Stoeckli, S.J.; Moch, H. Podoplanin expression correlates with sentinel lymph node metastasis in early squamous cell carcinomas of the oral cavity and oropharynx. Int. J. Cancer 2010, 129, 1404–1409.
- Kilvaer, T.K.; Valkov, A.; Sorbye, S.; Smeland, E.; Bremnes, R.M.; Busund, L.-T.; Donnem, T. Profiling of VEGFs and VEGFRs as Prognostic Factors in Soft Tissue Sarcoma: VEGFR-3 Is an Independent Predictor of Poor Prognosis. PLoS ONE 2010, 5, e15368.
- Liu, B.; Ma, J.; Wang, X.; Su, F.; Li, X.; Yang, S.; Ma, W.; Zhang, Y. Lymphangiogenesis and Its Relationship With Lymphatic Metastasis and Prognosis in Malignant Melanoma. Anat. Rec. 2008, 291, 1227–1235.
- Buzrla, P.; Dvorackova, J.; Motyka, O. Lymphangiogenesis and Its Correlation with the VEGF Expression and the Sentinel Lymph Node in Cutaneous Melanomas. BioMed Res. Int. 2014, 2014, 372979.
- Johnson, K.E.; Wilgus, T.A. Multiple Roles for VEGF in Non-Melanoma Skin Cancer: Angiogenesis and Beyond. J. Ski. Cancer 2012, 2012, 483439.
- Gallego, E.; Vicioso, L.; Alvarez, M.; Hierro, I.; Perez-Villa, L.; Blanes, A.; Matilla, A. Stromal expression of vascular endo- thelial growth factor C is relevant to predict sentinel lymph node status in melanomas. Virchows Arch. 2011, 458, 621–630.
- Boone, B.; Blokx, W.; De Bacquer, D.; Lambert, J.; Ruiter, D.; Brochez, L. The role of VEGF-C staining in predicting regional metastasis in melanoma. Virchows Arch. 2008, 453, 257–265.
- Brychtova, S.; Bezdekova, M.; Brychta, T.; Tichy, M. The role of vascular endothelial growth factors and their receptors in malignant melanomas. Neoplasma 2008, 55, 273–279.
- Dadras, S.S.; Lange-Asschenfeldt, B.; Velasco, P.; Nguyen, L.; Vora, A.; Muzikansky, A.; Jahnke, K.; Hauschild, A.; Hirakawa, S.; Mihm, M.C.; et al. Tumor lymphangiogenesis predicts melanoma metastasis to sentinel lymph nodes. Mod. Pathol. 2005, 18, 1232–1242.
- Toberer, F.; Haenssle, H.; Laimer, M.; Heinzel-Gutenbrunner, M.; Enk, A.; Hartschuh, W.; Helmbold, P.; Kutzner, H. Vascular Endothelial Growth Factor Receptor-3 Expression Predicts Sentinel Node Status in Primary Cutaneous Melanoma. Acta Derm. Venereol. 2020, 100, adv00235.
- Hemler, M.E. Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 2013, 14, 49–60.
- Fan, J.; Zhu, G.-Z.; Niles, R.M. Expression and function of CD9 in melanoma cells. Mol. Carcinog. 2009, 49, 85–93.
- Powner, D.; Kopp, P.M.; Monkley, S.J.; Critchley, D.R.; Berditchevski, F. Tetraspanin CD9 in cell migration. Biochem. Soc. Trans. 2011, 39, 563–567.
- Lucarini, G.; Molinelli, E.; Licini, C.; Rizzetto, G.; Radi, G.; Goteri, G.; Mattioli-Belmonte, M.; Offidani, A.; Simonetti, O. Tetraspanin CD9 Expression Predicts Sentinel Node Status in Patients with Cutaneous Melanoma. Int. J. Mol. Sci. 2022, 23, 4775.
- Liang, P.; Miao, M.; Liu, Z.; Wang, H.; Jiang, W.; Ma, S.; Li, C.; Hu, R. CD9 expression indicates a poor outcome in acute lymphoblastic leukemia. Cancer Biomark. 2018, 21, 781–786.
- Huan, J.; Gao, Y.; Xu, J.; Sheng, W.; Zhu, W.; Zhang, S.; Cao, J.; Ji, J.; Zhang, L.; Tian, Y. Overexpression of CD9 correlates with tumor stage and lymph node metastasis in esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3054–3061.
- Tasdemir, A.; Soyuer, I.; Unal, D.; Artis, T. Prognostic value of NF-κB, CD9, and VEGF in gastrointestinal stromal tumors. Contemp. Oncol. 2013, 17, 493–498.
- Miki, Y.; Yashiro, M.; Okuno, T.; Kitayama, K.; Masuda, G.; Hirakawa, K.; Ohira, M. CD9-positive exosomes from cancer-associated fibroblasts stimulate the migration ability of scirrhous-type gastric cancer cells. Br. J. Cancer 2018, 118, 867–877.
- Lu, W.; Fei, A.; Jiang, Y.; Chen, L.; Wang, Y. Tetraspanin CD9 interacts with α-secretase to enhance its oncogenic function in pancreatic cancer. Am. J. Transl. Res. 2020, 12, 5525–5537.
- Nagare, R.P.; Sneha, S.; Krishnapriya, S.; Ramachandran, B.; Murhekar, K.; Vasudevan, S.; Shabna, A.; Ganesan, T.S. ALDH1A1+ ovarian cancer stem cells co-expressing surface markers CD24, EPHA1 and CD9 form tumours in vivo. Exp. Cell Res. 2020, 392, 112009.
- Logozzi, M.; Di Raimo, R.; Mizzoni, D.; Fais, S. Immunocapture-based ELISA to characterize and quantify exosomes in both cell culture supernatants and body fluids. Methods Enzymol. 2020, 645, 155–180.
- Longo, N.; Yáñez-Mó, M.; Mittelbrunn, M.; De La Rosa, G.; Muňoz, M.-L.; Sánchez-Madrid, F.; Sánchez-Mateos, P. Regulatory role of tetraspanin CD9 in tumor–endothelial cell interaction during transendothelial invasion of melanoma cells. Blood 2001, 98, 3717–3726.
- Erovic, B.M.; Neuchrist, C.; Kandutsch, S.; Woegerbauer, M.; Pammer, J. CD9 Expression on Lymphatic Vessels in Head and Neck Mucosa. Mod. Pathol. 2003, 16, 1028–1034.
- Doeden, K.; Ma, Z.; Narasimhan, B.; Swetter, S.M.; Detmar, M.; Dadras, S.S. Lymphatic invasion in cutaneous melanoma is associated with sentinel lymph node metastasis. J. Cutan. Pathol. 2009, 36, 772–780.
- Fohn, L.E.; Rodriguez, A.; Kelley, M.C.; Ye, F.; Shyr, Y.; Stricklin, G.; Robbins, J.B. D2-40 lymphatic marker for detecting lymphatic invasion in thin to intermediate thickness melanomas: Association with sentinel lymph node status and prognostic value—A retrospective case study. J. Am. Acad. Dermatol. 2011, 64, 336–345.
- Gerami, P.; Cook, R.W.; Russell, M.C.; Wilkinson, J.; Amaria, R.N.; Gonzalez, R.; Lyle, S.; Jackson, G.L.; Greisinger, A.J.; Johnson, C.E.; et al. Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J. Am. Acad. Dermatol. 2015, 72, 780–785.e3.
- Cook, R.W.; Middlebrook, B.; Wilkinson, J.; Covington, K.R.; Oelschlager, K.; Monzon, F.A.; Stone, J.F. Analytic validity of DecisionDx-Melanoma, a gene expression profile test for determining metastatic risk in melanoma patients. Diagn. Pathol. 2018, 13, 13.
- Keller, J.; Schwartz, T.L.; Lizalek, J.M.; Chang, E.; Patel, A.D.; Hurley, M.Y.; Hsueh, E.C. Prospective validation of the prognostic 31-gene expression profiling test in primary cutaneous melanoma. Cancer Med. 2019, 8, 2205–2212.
- Podlipnik, S.; Carrera, C.; Boada, A.; Richarz, N.A.; López-Estebaranz, J.; Pinedo-Moraleda, F.; Elosua-González, M.; Martín-González, M.; Carrillo-Gijón, R.; Redondo, P.; et al. Early outcome of a 31-gene expression profile test in 86 AJCC stage IB–II melanoma patients. A prospective multicentre cohort study. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 857–862.
- Greenhaw, B.N.; Zitelli, J.A.; Brodland, D.G. Estimation of Prognosis in Invasive Cutaneous Melanoma: An Independent Study of the Accuracy of a Gene Expression Profile Test. Dermatol. Surg. 2018, 44, 1494–1500.
- Hsueh, E.C.; DeBloom, J.R.; Lee, J.; Sussman, J.J.; Covington, K.R.; Middlebrook, B.; Johnson, C.; Cook, R.W.; Slingluff, C.L., Jr.; McMasters, K.M. Interim analysis of survival in a prospective, multi-center registry cohort of cutaneous melanoma tested with a prognostic 31-gene expression profile test. J. Hematol. Oncol. 2017, 10, 152.
- Vetto, J.T.; Hsueh, E.C.; Gastman, B.R.; Dillon, L.D.; Monzon, F.A.; Cook, R.W.; Keller, J.; Huang, X.; Fleming, A.; Hewgley, P.; et al. Guidance of sentinel lymph node biopsy decisions in patients with T1–T2 melanoma using gene expression profiling. Futur. Oncol. 2019, 15, 1207–1217.
- Gastman, B.R.; Gerami, P.; Kurley, S.J.; Cook, R.W.; Leachman, S.; Vetto, J.T. Identification of patients at risk of metastasis using a prognostic 31-gene expression profile in subpopulations of melanoma patients with favorable outcomes by standard criteria. J. Am. Acad. Dermatol. 2018, 80, 149–157.e4.
- Carr, M.J.; Monzon, F.A.; Zager, J.S. Sentinel lymph node biopsy in melanoma: Beyond histologic factors. Clin. Exp. Metastasis 2021, 39, 29–38.
More
Information
Subjects:
Dermatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
796
Revisions:
2 times
(View History)
Update Date:
03 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




