Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mario Piccioli | -- | 1845 | 2022-12-02 13:15:00 | | | |
| 2 | Camila Xu | Meta information modification | 1845 | 2022-12-05 00:59:49 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Camponeschi, F.; Piccioli, M.; Banci, L. The Peculiar Properties of Human mitoNEET. Encyclopedia. Available online: https://encyclopedia.pub/entry/37881 (accessed on 07 February 2026).
Camponeschi F, Piccioli M, Banci L. The Peculiar Properties of Human mitoNEET. Encyclopedia. Available at: https://encyclopedia.pub/entry/37881. Accessed February 07, 2026.
Camponeschi, Francesca, Mario Piccioli, Lucia Banci. "The Peculiar Properties of Human mitoNEET" Encyclopedia, https://encyclopedia.pub/entry/37881 (accessed February 07, 2026).
Camponeschi, F., Piccioli, M., & Banci, L. (2022, December 02). The Peculiar Properties of Human mitoNEET. In Encyclopedia. https://encyclopedia.pub/entry/37881
Camponeschi, Francesca, et al. "The Peculiar Properties of Human mitoNEET." Encyclopedia. Web. 02 December, 2022.
Copy Citation
The outer mitochondrial membrane (OMM) protein mitoNEET, also known as CDGSH Fe-S domain-containing protein-1 (CISD1), is composed of 108 amino acids, encompassing a N-terminal transmembrane helix (residues 14–32) that anchors the protein to the OMM, and a cytosolic portion (residues 33–108) that has been widely investigated through X-ray crystallography, showing a unique, highly conserved folding.
iron-sulfur proteins
paramagnetic NMR
iron-sulfur cluster biogenesis
1. Introduction
Proteins require specific folds and three-dimensional structures in order to perform specific functions; hence, the mutation or misfolding of proteins are often associated with serious diseases. For proteins that bind cofactors, the latter play fundamental roles either in ensuring the correct three-dimensional folding of the protein, or being directly involved in the biological function of the protein itself. Fe-S clusters are ancient cofactors that are involved in a multiplicity of functions, such as electron transfer, metabolic reactions, gene expression, regulation, and DNA maintenance [1][2][3][4][5][6]. Many factors determine the wide range of specific characters taken by each Fe-S cluster: their redox potential can be tuned by the protein environment over almost 1 V range; single or multiple cluster(s) can be accommodated in a protein, either with identical or different functions; many consensus sequences are able to bind Fe-S cluster, thus providing different topologies. The functional diversity of Fe-S proteins is, therefore, strongly correlated to the properties of the cluster(s) and its neighbor region. Due to these features, Fe-S proteins have been, for four decades, a playground for bio-spectroscopists of different kinds. Recently, a novel family of [2Fe-2S] proteins, called “NEET” proteins due to the presence of the Asn-Glu-Glu-Thr (NEET) amino acid sequence at their C-termini [7], has been discovered in several organisms [8], which has been found to be involved in many processes related to normal cellular metabolism and diseases [9]. In humans, three NEET proteins are encoded by the CISD1, CISD2, and CISD3 genes. The three proteins share common structural, biochemical, and spectroscopic features, as well as similar cellular roles in the regulation of iron and ROS homeostasis in cells [9][10].
2. The Peculiar Properties of Human mitoNEET: A Unique Folding for a Multiplicity of Functions
The outer mitochondrial membrane (OMM) protein mitoNEET, also known as CDGSH Fe-S domain-containing protein-1 (CISD1), is composed of 108 amino acids, encompassing a N-terminal transmembrane helix (residues 14–32) that anchors the protein to the OMM [11], and a cytosolic portion (residues 33–108) that has been widely investigated through X-ray crystallography, showing a unique, highly conserved folding [12][13][14][15]. All the crystallographic structures of mitoNEET revealed the presence of two distinct domains: a β-rich or “β-cap” region and a cluster-binding domain (Figure 1a) [12][13][15][16]. The latter contains the highly conserved CXCX2(S/T)X3PXCDG(S/A/T)H motif that binds a [2Fe-2S] cluster, and that is part of the characteristic CDGSH-type domain of 39 amino acids, which constitutes the hallmark of the “NEET” family.
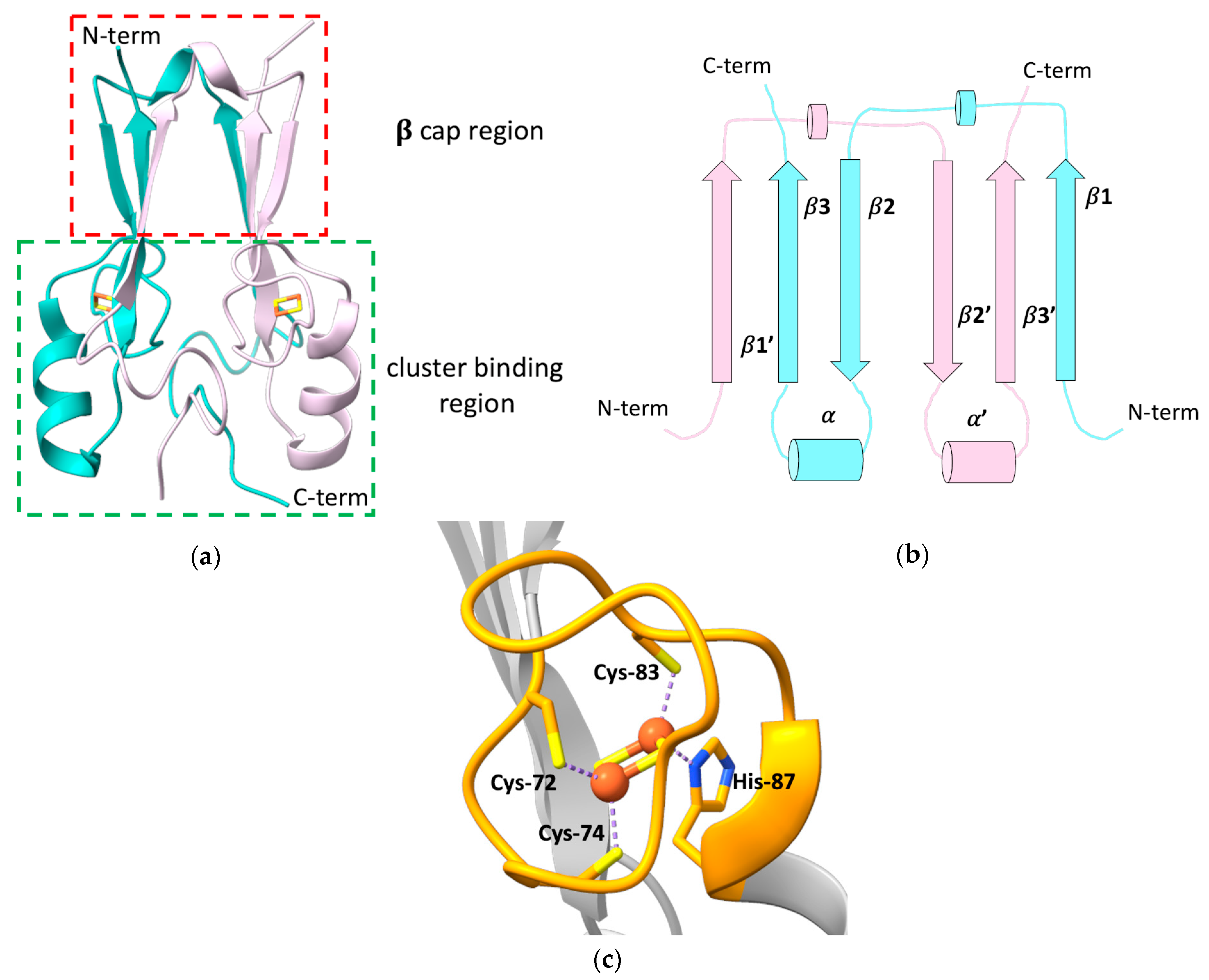
Figure 1. (a) Crystallographic structure of the soluble domain of human mitoNEET, with highlighted the β-cap and the cluster-binding regions (PDB ID: 2QH7). The two subunits forming the dimer are reported in different colors; (b) topology diagram illustrating the organization of the secondary structural units in the two protomers of the dimeric structure of mitoNEET; (c) [2Fe-2S] cluster-binding motif of human mitoNEET. The conserved CXCX2(S/T)X3PXCDG(S/A/T)H motif is highlighted in orange.
mitoNEET forms an intertwined, parallel homodimer, with a pseudo two-fold symmetry [12][13][16]. Each protomer of the dimer comprises a long, flexible loop in the N-terminal part (res 33–55), whose structure has not been solved yet [14], and displays a β1-β2-α-β4 topology in the rest of the protein (Figure 1b).
In the dimer, the β-strands form two symmetric β-sheets, composed of two antiparallel β-strands from one monomer (residues 68–71 and 101–104) and an additional parallel swapped-strand from the other monomer (residues 56–61) [12][13][15]. These two β-sheets and the two loops connecting the swapped β-strands and also containing a helical turn (residues 62–64), form the so-called β-cap region [12][13][15][16], whereas the N-terminal portion of the soluble domain, the conserved α-helix formed by residues 86–94 and the loop connecting the helix to the β-cap, together with the CXCX2(S/T)X3PXCDG(S/A/T)H motif, form the cluster-binding domain [12][13][15][16]. Here, two symmetric hydrophobic cores, comprising Ile-45, Ile-56, Trp-75, Phe-80 of one monomer, and Val-98 of the second monomer [13], and two intermolecular hydrogen bonds between His-58 and Arg-73 [15], further stabilize the dimeric state of mitoNEET.
The folding of the two domains is indeed interdependent. Theoretical structure-based folding studies proposed that the rigidity in the β-cap region creates a constraint for the folding of the cluster-binding domain [16]. As part of the folding process, mitoNEET binds two [2Fe-2S]2+/+ clusters, one in each subunit of the dimer, using three cysteines (Cys-72, Cys-74, Cys-83) and one histidine (His-87) as ligands (Figure 1c). The His-87 ligand is located at the N-terminus of the α-helix within the cluster-binding domain and is solvent-accessible, as it is the Fe ion that it coordinates together with Cys-83. The other two ligands (Cys-72 and Cys-74) are, by contrast, buried inside the structure and bind a non-solvent-accessible Fe ion [14][15][17]. This coordination sphere, that is common to all the NEET proteins, is different from those of ferredoxin-like and Rieske proteins that coordinate [2Fe-2S]2+/+ clusters either with four cysteines, or with two cysteines and two histidines, respectively. The binding of the [2Fe-2S] clusters to each subunit in the dimer is responsible for the unique fold adopted by the holo protein [18], and the dimeric arrangement of the CDGSH domains seems to be essential for the stabilization of the coordination sphere of mitoNEET. Indeed it has been proposed that the β-cap domain formed by the strand swapping of the two protomers could function as an allosteric control site, modulating cluster insertion, assembly, or electron transfer [16]. Even CISD3, considered as the ancestor protein of the family, being monomeric, contains two CDGSH domains in its primary sequence and, as a consequence, two [2Fe-2S] clusters [19]. No cooperativity effect between the two clusters has been reported so far, even though inter-cluster dipolar coupling has been detected by EPR studies [20].
MitoNEET takes part into a variety of cellular processes, acting as a regulator of the homeostasis of iron and of reactive oxygen species (ROS) [21][22], as well as of the metabolism of glucose and lipids in cells, therefore modulating mitochondrial bioenergetics [21][23][24]. These functions have been linked to the observed overexpression of mitoNEET in human epithelial breast cancer cells, where the protein has been found to induce tumor cell proliferation, likely maintaining the mitochondrial functions by preventing the accumulation of iron and ROS in the mitochondrial matrix, regulating autophagy signaling, preventing autophagy [25][26][27]. Recent studies reported that mitoNEET indeed controls the formation and the integrity of inter-mitochondrial junctions and mitochondrial network morphology [24][28]. Moreover, it was observed that mitoNEET interacts with the voltage-dependent anion channel 1 (VDAC), thus regulating the free iron level within mitochondria [29]. This interaction was also proposed to be functional for the maturation of mitoNEET clusters, mediating the interaction between mitoNEET and CISD3 in a process where CISD3 transfers its [2Fe-2S] clusters from inside the mitochondria to mitoNEET [30]. In addition, mitoNEET was found to be involved in several human pathologies, such as obesity, where the overexpression of mitoNEET enhances lipid accumulation in adipocytes, but preserves insulin sensitivity [21][31][32] and neurodegeneration [33]. MitoNEET have been also linked to type 2 diabetes, since it was identified as the main cellular target of the thiazolidinedione (TZD) pioglitazone, a drug extensively used to treat insulin resistance [7], although the role of mitoNEET in the etiology of the pathology is unclear.
Despite the number of cellular and pathological mitoNEET-related processes, very few details are known about the mechanism of action of the protein in such processes. MitoNEET is supposed to play a major role in repairing the damaged [4Fe-4S] cluster on cytosolic apo aconitase IRP1 in oxidative stress conditions [18], and acts as a cluster transfer protein for several apo recipient proteins [18][34][35]. Both functions are based on a redox switch, activated by several cellular components [36][37][38][39][40]. Indeed, only [2Fe-2S]2+ and not [2Fe-2S]+ clusters can be transferred from holo mitoNEET to apo recipient proteins [18][34][35]. For this reason, the oxidized and the reduced state of mitoNEET clusters have been defined as “active” and “dormant” states, respectively [41]. The two [2Fe-2S] clusters of mitoNEET have a ∼0 mV midpoint redox potential in vitro at pH 7.5 [42][43] and in the cytoplasmic cellular environment, they are in the reduced state, as shown by EPR spectroscopy performed on E. coli cells containing the overexpressed cytosolic domain of human mitoNEET [44]. These findings suggest that in normal cellular conditions, mitoNEET clusters are stably bound to the protein in the reduced, dormant state. Several factors can change the redox state, and, therefore, the reactivity of mitoNEET clusters: [2Fe-2S]2+-mitoNEET can be reduced in vitro by biological thiols [44], reduced flavin nucleotides [36][37][38], and enzymes, such as human glutathione reductase [39]. Additionally, other Fe-S proteins, such as human anamorsin, transfer electrons to mitoNEET in vitro, showing a possible direct link between the cytosolic iron-sulfur cluster assembly (CIA) machinery and the mitoNEET cluster transfer repairing pathway [40].
Another important factor for the stability/reactivity of mitoNEET is the peculiar pH lability of its [2Fe-2S] clusters. The presence of a His residue in the first coordination sphere of mitoNEET [2Fe-2S] clusters results in a significant sensitivity to pH variations and in the pH lability of the cluster, described as a peculiar feature of the NEET proteins. Indeed, it has been proposed that the protonation of the imidazolic ring of the His-87 ligand at acidic pH facilitates the transfer of the mitoNEET [2Fe-2S]2+ clusters to apo recipient proteins in vitro, or their release in solution [18][34][35][41]. However, the protonation of the His-87 ligand is likely not the sole factor affecting cluster stability in mitoNEET. Indeed, Rieske proteins, which contain two His residues in their cluster coordination sphere, show a significant higher cluster stability over a wide range of pH [45]. The pH-dependent stability of mitoNEET clusters seems to be related also to a hydrogen-bonding network formed by the His-87 ligand, a conserved solvent water molecule, and the Nε of Lys-55 residue from the other polypeptide chain of the dimer [42][46][47]. Moreover, NMR and UV-visible spectroscopies showed that the stability of mitoNEET [2Fe-2S] clusters can be tuned also by the interaction with small molecules, such as the antidiabetic drug pioglitazone, which interacts with the cluster-binding region of the protein, increasing the stability of the two Fe-S clusters [15][48], or reduced nicotinamide adenine dinucleotide phosphate (NADPH), which, by contrast, destabilizes mitoNEET Fe-S clusters and induces protein unfolding [49].
For all the aforementioned characteristics, the study of the electronic and coordination structures of mitoNEET [2Fe-2S] clusters has been attracting increasing interest in the last few years, and, in addition to the above-described X-ray crystallographic studies, a variety of spectroscopic techniques have been applied to study mitoNEET in the attempt to explain the drastic difference in clusters stability and reactivity observed for the two redox states and to clarify the cellular function of the protein. The similarities and the differences between the oxidized and reduced states of the mitoNEET [2Fe-2S] clusters and those of other Fe-S proteins are crucial aspects for understanding their modes of action and their role in the physiological processes.
References
- Beinert, H. Iron-Sulfur Proteins: Ancient Structures, Still Full of Surprises. J. Biol. Inorg. Chem. 2000, 5, 2–15.
- Lill, R. Function and Biogenesis of Iron-Sulphur Proteins. Nature 2009, 460, 831–838.
- Rouault, T.A. The Indispensable Role of Mammalian Iron Sulfur Proteins in Function and Regulation of Multiple Diverse Metabolic Pathways. Biometals 2019, 32, 343–353.
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, Function, and Formation of Biological Iron-Sulfur Clusters. Annu. Rev. Biochem. 2005, 74, 247–281.
- Golinelli-Cohen, M.-P.; Bouton, C. Fe-S Proteins Acting as Redox Switch: New Key Actors of Cellular Adaptive Responses. Curr. Chem. Biol. 2017, 11, 70–88.
- Fuss, J.O.; Tsai, C.-L.; Ishida, J.P.; Tainer, J.A. Emerging Critical Roles of Fe-S Clusters in DNA Replication and Repair. Biochim. Biophys. Acta 2015, 1853, 1253–1271.
- Colca, J.R.; McDonald, W.G.; Waldon, D.J.; Leone, J.W.; Lull, J.M.; Bannow, C.A.; Lund, E.T.; Mathews, W.R. Identification of a Novel Mitochondrial Protein (“mitoNEET”) Cross-Linked Specifically by a Thiazolidinedione Photoprobe. Am. J. Physiol. Endocrinol. Metab. 2004, 286, E252–E260.
- Lin, J.; Zhang, L.; Lai, S.; Ye, K. Structure and Molecular Evolution of CDGSH Iron-Sulfur Domains. PLoS ONE 2011, 6, e24790.
- Mittler, R.; Darash-Yahana, M.; Sohn, Y.S.; Bai, F.; Song, L.; Cabantchik, I.Z.; Jennings, P.A.; Onuchic, J.N.; Nechushtai, R. NEET Proteins: A New Link Between Iron Metabolism, Reactive Oxygen Species, and Cancer. Antioxid. Redox Signal. 2019, 30, 1083–1095.
- Nechushtai, R.; Karmi, O.; Zuo, K.; Marjault, H.-B.; Darash-Yahana, M.; Sohn, Y.-S.; King, S.D.; Zandalinas, S.I.; Carloni, P.; Mittler, R. The Balancing Act of NEET Proteins: Iron, ROS, Calcium and Metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118805.
- Wiley, S.E.; Murphy, A.N.; Ross, S.A.; van der Geer, P.; Dixon, J.E. MitoNEET Is an Iron-Containing Outer Mitochondrial Membrane Protein That Regulates Oxidative Capacity. Proc. Natl. Acad. Sci. USA 2007, 104, 5318–5323.
- Hou, X.; Liu, R.; Ross, S.; Smart, E.J.; Zhu, H.; Gong, W. Crystallographic Studies of Human MitoNEET. J. Biol. Chem. 2007, 282, 33242–33246.
- Lin, J.; Zhou, T.; Ye, K.; Wang, J. Crystal Structure of Human MitoNEET Reveals Distinct Groups of Iron–Sulfur Proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 14640–14645.
- Conlan, A.R.; Paddock, M.L.; Axelrod, H.L.; Cohen, A.E.; Abresch, E.C.; Wiley, S.; Roy, M.; Nechushtai, R.; Jennings, P.A. The Novel 2Fe–2S Outer Mitochondrial Protein MitoNEET Displays Conformational Flexibility in Its N-Terminal Cytoplasmic Tethering Domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 654–659.
- Paddock, M.L.; Wiley, S.E.; Axelrod, H.L.; Cohen, A.E.; Roy, M.; Abresch, E.C.; Capraro, D.; Murphy, A.N.; Nechushtai, R.; Dixon, J.E.; et al. MitoNEET Is a Uniquely Folded 2Fe 2S Outer Mitochondrial Membrane Protein Stabilized by Pioglitazone. Proc. Natl. Acad. Sci. USA 2007, 104, 14342–14347.
- Baxter, E.L.; Jennings, P.A.; Onuchic, J.N. Interdomain Communication Revealed in the Diabetes Drug Target MitoNEET. Proc. Natl. Acad. Sci. USA 2011, 108, 5266–5271.
- Tamir, S.; Paddock, M.L.; Darash-Yahana-Baram, M.; Holt, S.H.; Sohn, Y.S.; Agranat, L.; Michaeli, D.; Stofleth, J.T.; Lipper, C.H.; Morcos, F.; et al. Structure–Function Analysis of NEET Proteins Uncovers Their Role as Key Regulators of Iron and ROS Homeostasis in Health and Disease. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 1294–1315.
- Ferecatu, I.; Gonçalves, S.; Golinelli-Cohen, M.-P.; Clémancey, M.; Martelli, A.; Riquier, S.; Guittet, E.; Latour, J.-M.; Puccio, H.; Drapier, J.-C.; et al. The Diabetes Drug Target MitoNEET Governs a Novel Trafficking Pathway to Rebuild an Fe-S Cluster into Cytosolic Aconitase/Iron Regulatory Protein 1. J. Biol. Chem. 2014, 289, 28070–28086.
- Lipper, C.H.; Karmi, O.; Sohn, Y.S.; Darash-Yahana, M.; Lammert, H.; Song, L.; Liu, A.; Mittler, R.; Nechushtai, R.; Onuchic, J.N.; et al. Structure of the Human Monomeric NEET Protein MiNT and Its Role in Regulating Iron and Reactive Oxygen Species in Cancer Cells. Proc. Natl. Acad. Sci. USA 2018, 115, 272–277.
- Iwasaki, T.; Samoilova, R.I.; Kounosu, A.; Ohmori, D.; Dikanov, S.A. Continuous-Wave and Pulsed EPR Characterization of the (Cys)3(His)1 Cluster in Rat MitoNEET. J. Am. Chem. Soc. 2009, 131, 13659–13667.
- Kusminski, C.M.; Holland, W.L.; Sun, K.; Park, J.; Spurgin, S.B.; Lin, Y.; Askew, G.R.; Simcox, J.A.; McClain, D.A.; Li, C.; et al. MitoNEET-Driven Alterations in Adipocyte Mitochondrial Activity Reveal a Crucial Adaptive Process That Preserves Insulin Sensitivity in Obesity. Nat. Med. 2012, 18, 1539–1549.
- Lee, S.; Seok, B.G.; Lee, S.-J.; Chung, S.W. Inhibition of MitoNEET Attenuates LPS-Induced Inflammation and Oxidative Stress. Cell Death Dis. 2022, 13, 127.
- Yonutas, H.M.; Hubbard, W.B.; Pandya, J.D.; Vekaria, H.J.; Geldenhuys, W.J.; Sullivan, P.G. Bioenergetic Restoration and Neuroprotection after Therapeutic Targeting of MitoNEET: New Mechanism of Pioglitazone Following Traumatic Brain Injury. Exp. Neurol. 2020, 327, 113243.
- Vernay, A.; Marchetti, A.; Sabra, A.; Jauslin, T.N.; Rosselin, M.; Scherer, P.E.; Demaurex, N.; Orci, L.; Cosson, P. MitoNEET-Dependent Formation of Intermitochondrial Junctions. Proc. Natl. Acad. Sci. USA 2017, 114, 8277–8282.
- Sohn, Y.-S.; Tamir, S.; Song, L.; Michaeli, D.; Matouk, I.; Conlan, A.R.; Harir, Y.; Holt, S.H.; Shulaev, V.; Paddock, M.L.; et al. NAF-1 and MitoNEET Are Central to Human Breast Cancer Proliferation by Maintaining Mitochondrial Homeostasis and Promoting Tumor Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 14676–14681.
- Salem, A.F.; Whitaker-Menezes, D.; Howell, A.; Sotgia, F.; Lisanti, M.P. Mitochondrial Biogenesis in Epithelial Cancer Cells Promotes Breast Cancer Tumor Growth and Confers Autophagy Resistance. Cell Cycle 2012, 11, 4174–4180.
- Geldenhuys, W.J.; Piktel, D.; Moore, J.C.; Rellick, S.L.; Meadows, E.; Pinti, M.V.; Hollander, J.M.; Ammer, A.G.; Martin, K.H.; Gibson, L.F. Loss of the Redox Mitochondrial Protein MitoNEET Leads to Mitochondrial Dysfunction in B-Cell Acute Lymphoblastic Leukemia. Free Radic. Biol. Med. 2021, 175, 226–235.
- Molino, D.; Pila-Castellanos, I.; Marjault, H.-B.; Dias Amoedo, N.; Kopp, K.; Rochin, L.; Karmi, O.; Sohn, Y.-S.; Lines, L.; Hamaï, A.; et al. Chemical Targeting of NEET Proteins Reveals Their Function in Mitochondrial Morphodynamics. EMBO Rep. 2020, 21, e49019.
- Lipper, C.H.; Stofleth, J.T.; Bai, F.; Sohn, Y.-S.; Roy, S.; Mittler, R.; Nechushtai, R.; Onuchic, J.N.; Jennings, P.A. Redox-Dependent Gating of VDAC by MitoNEET. Proc. Natl. Acad. Sci. USA 2019, 116, 19924–19929.
- Karmi, O.; Marjault, H.-B.; Bai, F.; Roy, S.; Sohn, Y.-S.; Darash Yahana, M.; Morcos, F.; Ioannidis, K.; Nahmias, Y.; Jennings, P.A.; et al. A VDAC1-Mediated NEET Protein Chain Transfers Clusters between the Mitochondria and the Cytosol and Impacts Mitochondrial Dynamics. Proc. Natl. Acad. Sci. USA 2022, 119, e2121491119.
- Kusminski, C.M.; Park, J.; Scherer, P.E. MitoNEET-Mediated Effects on Browning of White Adipose Tissue. Nat. Commun. 2014, 5, 3962.
- Moreno-Navarrete, J.M.; Moreno, M.; Ortega, F.; Sabater, M.; Xifra, G.; Ricart, W.; Fernández-Real, J.M. CISD1 in Association with Obesity-Associated Dysfunctional Adipogenesis in Human Visceral Adipose Tissue. Obesity 2016, 24, 139–147.
- Geldenhuys, W.J.; Benkovic, S.A.; Lin, L.; Yonutas, H.M.; Crish, S.D.; Sullivan, P.G.; Darvesh, A.S.; Brown, C.M.; Richardson, J.R. MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson’s Disease Phenotype. ACS Chem. Neurosci. 2017, 8, 2759–2765.
- Lipper, C.H.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Nechushtai, R.; Jennings, P.A. Cancer-Related NEET Proteins Transfer 2Fe-2S Clusters to Anamorsin, a Protein Required for Cytosolic Iron-Sulfur Cluster Biogenesis. PLoS ONE 2015, 10, e0139699.
- Zuris, J.A.; Harir, Y.; Conlan, A.R.; Shvartsman, M.; Michaeli, D.; Tamir, S.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Cabantchik, Z.I.; et al. Facile Transfer of Clusters from the Diabetes Drug Target MitoNEET to an Apo-Acceptor Protein. Proc. Natl. Acad. Sci. USA 2011, 108, 13047–13052.
- Landry, A.P.; Wang, Y.; Cheng, Z.; Crochet, R.B.; Lee, Y.-H.; Ding, H. Flavin Nucleotides Act as Electron Shuttles Mediating Reduction of the Clusters in Mitochondrial Outer Membrane Protein MitoNEET. Free Radic. Biol. Med. 2017, 102, 240–247.
- Wang, Y.; Landry, A.P.; Ding, H. The Mitochondrial Outer Membrane Protein MitoNEET Is a Redox Enzyme Catalyzing Electron Transfer from FMNH2 to Oxygen or Ubiquinone. J. Biol. Chem. 2017, 292, 10061–10067.
- Tasnim, H.; Landry, A.P.; Fontenot, C.R.; Ding, H. Exploring the FMN Binding Site in the Mitochondrial Outer Membrane Protein MitoNEET. Free Radic. Biol. Med. 2020, 156, 11–19.
- Landry, A.P.; Cheng, Z.; Ding, H. Reduction of Mitochondrial Protein MitoNEET Clusters by Human Glutathione Reductase. Free Radic. Biol. Med. 2015, 81, 119–127.
- Camponeschi, F.; Ciofi-Baffoni, S.; Banci, L. Anamorsin/Ndor1 Complex Reduces -MitoNEET via a Transient Protein-Protein Interaction. J. Am. Chem. Soc. 2017, 139, 9479–9482.
- Golinelli-Cohen, M.-P.; Lescop, E.; Mons, C.; Gonçalves, S.; Clémancey, M.; Santolini, J.; Guittet, E.; Blondin, G.; Latour, J.-M.; Bouton, C. Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster. J. Biol. Chem. 2016, 291, 7583–7593.
- Bak, D.W.; Zuris, J.A.; Paddock, M.L.; Jennings, P.A.; Elliott, S.J. Redox Characterization of the FeS Protein MitoNEET and Impact of Thiazolidinedione Drug Binding. Biochemistry 2009, 48, 10193–10195.
- Tirrell, T.F.; Paddock, M.L.; Conlan, A.R.; Smoll, E.J.; Nechushtai, R.; Jennings, P.A.; Kim, J.E. Resonance Raman Studies of the (His)(Cys)3 2Fe-2S Cluster of MitoNEET: Comparison to the (Cys)4 Mutant and Implications of the Effects of PH on the Labile Metal Center. Biochemistry 2009, 48, 4747–4752.
- Landry, A.P.; Ding, H. Redox Control of Human Mitochondrial Outer Membrane Protein MitoNEET Clusters by Biological Thiols and Hydrogen Peroxide. J. Biol. Chem. 2014, 289, 4307–4315.
- Schröter, T.; Hatzfeld, O.M.; Gemeinhardt, S.; Korn, M.; Friedrich, T.; Ludwig, B.; Link, T.A. Mutational Analysis of Residues Forming Hydrogen Bonds in the Rieske Cluster of the Cytochrome Bc1 Complex in Paracoccus Denitrificans. Eur. J. Biochem. 1998, 255, 100–106.
- Bak, D.W.; Elliott, S.J. Conserved Hydrogen Bonding Networks of MitoNEET Tune FeS Cluster Binding and Structural Stability. Biochemistry 2013, 52, 4687–4696.
- Pesce, L.; Calandrini, V.; Marjault, H.-B.; Lipper, C.H.; Rossetti, G.; Mittler, R.; Jennings, P.A.; Bauer, A.; Nechushtai, R.; Carloni, P. Molecular Dynamics Simulations of the Cluster-Binding Domain of NEET Proteins Reveal Key Molecular Determinants That Induce Their Cluster Transfer/Release. J. Phys. Chem. B 2017, 121, 10648–10656.
- Song, G.; Tian, F.; Liu, H.; Li, G.; Zheng, P. Pioglitazone Inhibits Metal Cluster Transfer of MitoNEET by Stabilizing the Labile Fe-N Bond Revealed at Single-Bond Level. J. Phys. Chem. Lett. 2021, 12, 3860–3867.
- Zhou, T.; Lin, J.; Feng, Y.; Wang, J. Binding of Reduced Nicotinamide Adenine Dinucleotide Phosphate Destabilizes the Iron−Sulfur Clusters of Human MitoNEET. Biochemistry 2010, 49, 9604–9612.
More
Information
Subjects:
Biochemical Research Methods
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
816
Revisions:
2 times
(View History)
Update Date:
05 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




