Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ilona Jaszczuk | -- | 3059 | 2022-11-30 05:21:05 | | | |
| 2 | Sirius Huang | Meta information modification | 3059 | 2022-11-30 05:55:01 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jaszczuk, I.; Winkler, I.; Koczkodaj, D.; Skrzypczak, M.; Filip, A. The Role of Cluster C19MC in Pre-Eclampsia Development. Encyclopedia. Available online: https://encyclopedia.pub/entry/37241 (accessed on 07 February 2026).
Jaszczuk I, Winkler I, Koczkodaj D, Skrzypczak M, Filip A. The Role of Cluster C19MC in Pre-Eclampsia Development. Encyclopedia. Available at: https://encyclopedia.pub/entry/37241. Accessed February 07, 2026.
Jaszczuk, Ilona, Izabela Winkler, Dorota Koczkodaj, Maciej Skrzypczak, Agata Filip. "The Role of Cluster C19MC in Pre-Eclampsia Development" Encyclopedia, https://encyclopedia.pub/entry/37241 (accessed February 07, 2026).
Jaszczuk, I., Winkler, I., Koczkodaj, D., Skrzypczak, M., & Filip, A. (2022, November 30). The Role of Cluster C19MC in Pre-Eclampsia Development. In Encyclopedia. https://encyclopedia.pub/entry/37241
Jaszczuk, Ilona, et al. "The Role of Cluster C19MC in Pre-Eclampsia Development." Encyclopedia. Web. 30 November, 2022.
Copy Citation
Pre-eclampsia is a placenta-related complication occurring in 2–10% of all pregnancies. miRNAs are a group of non-coding RNAs regulating gene expression. There is evidence that C19MC miRNAs are involved in the development of the placenta.
pregnancy
pre-eclampsia
microRNAs
C19MC
1. The Chromosome 19 MicroRNA Cluster (C19MC)
The chromosome 19 microRNA cluster (C19MC) is a primate specific miRNA cluster located on the human chromosome 19q13.41 that is 100 kb in length. It contains 46 tandem repeating microRNA genes encoding 58 mature miRNAs [1][2]. C19MC is found only in primates and is almost exclusively expressed in the placenta, although low levels have also been shown in embryonic stem cells, testes and some tumors [3][4][5][6]. Many publications emphasize that the synthesis of miRNAs is highly orchestrated, and that in the placenta they are expressed at the appropriate time, in a tissue and species manner [7][8][9].
The level of C19MC miRNA expression in plasma and placenta increases with the advancing age of pregnancy, decreasing sharply after delivery [10][11]. In the plasma of pregnant women, C19MC miRNAs form a part of the placental-related fraction of circulating miRNAs or are packed into exosomes. The source of C19MC miRNAs in the plasma of pregnant women are cells of various areas of the placenta in which cells express C19MC (which can be studied after delivery), and also cells that during remodeling, undergo apoptosis, releasing placental debris and secreting exosomes into the maternal circulation [12][13].
Exosomes or nanovesicles are a small fraction (30–150 nm) of the extracellular vesicles (EVs) formed in multivesicular bodies (MVBs) [14] that are released by most cells into the extracellular space. Their function is to mediate intercellular communication through signaling molecules packed inside and secreted during exocytosis (proteins, lipids, RNA and DNA) after fusion with the cell membrane of target cells. Increased oxidative stress, observed in pre-eclampsia development, predisposes syncytiotrophoblast (STB) cells to the production of more humoral factors and to the release of microvesicles, including exosomes [15][16]. The composition of the miRNAs transported in exosomes in the PE is different compared to normal pregnancies [17]. Extracellular miRNAs packed into exosomes can be responsible for intercellular communication in an autocrine or paracrine manner, and at greater distances through circulation [18][19]. They can also modulate the immune response that ensures immune tolerance in the mother–fetus relationship or modify pro-inflammatory reactions in the course of pregnancy [20][21].
C19MC belongs to the genes encoding miRNAs whose expression is influenced by genomic imprinting, an epigenetic mechanism related to monoallelic expression in a parent-of-origin manner. C19MC is expressed exclusively from the paternal allele in the placenta which has been confirmed by single nucleotide polymorphism genotyping (SNP: G or T, rs55765443) mapping upstream the most 5′ microRNA transcribed by C19MC [22]. Many C19MC miRNAs are likely formed from the introns of a poorly characterized transcript called ”C19MC-HG”, composed of many repeating non-coding exons [1]. C19MC is expressed by the polymerase II promoter region, approximately 17 kb from the first exon, overlapping the differential methylation (DMR) region. The promoter is rich in CpG, showing the maternal characteristic methylation imprint acquired in the oocytes [22]. Maternal-specific methylation is deposited at CpG1, here termed ”C19MC-DMR1” (C19MC- differentially methylated region 1).
The structure and function of the C19MC cluster corresponds to features of imprinted genes present in the human genome. Imprinted genes form large chromosomal domains (up to 3 Mb), most of which are expressed in the placenta [23][24] and play an important role in prenatal embryo or placenta growth or regulate metabolic pathways in the placenta [25][26][27]. Regulation of imprinting gene expression in a given cluster is coordinated by epigenetically modified imprinting control regions (ICRs) that acquire a parental specific male DNA methylation imprint or female germline. In addition, a convergence has been observed between ICRs with maternal imprint and CpG-rich promoter regions [28]. DNA methylation at ICRs of imprinted genes is acquired during gametogenesis. Although methylation is a reversible process, the pattern of ICRs’ methylation is refractory to the genome-wide methylation reprogramming that occurs in the embryo after fertilization. DNA methylation levels can also be modified by the presence of specific SNPs [29], adjacent to the CpG islands in the in-cis system. It has been shown that C19MC miRNA transcription can be activated in cells by the use of DNA methylation inhibitors, confirming methylation-dependent epigenetic control in this region [22]. ICRs are themselves also marked by allele-specific post-translational histones modifications [2].
2. The Role of Cluster C19MC in Pre-Eclampsia Development
Chim et al., in 2008, were the first to highlight the potential use of miRNAs as biomarkers of pregnancy complications [30][31], while the chromosome 19 microRNA cluster (C19MC) was first described by Bentwich et al., in 2005 [32]. Noguer-Dance et al., in turn, showed that C19MC microRNAs were clearly expressed during embryo and placenta development. Moreover, they recognized that the imprinted C19MC miRNA genes had to evolve to improve the signaling pathways underlying primate morphology and placental development [17][22]. Their work also revealed that C19MC dysregulation leads to dysfunctional trophoblast cells, abnormal placentation and the consequent development of PE [17]. Zhang et al., in a subsequent study, showed that miR-515-5p was significantly decreased during the differentiation of human syncytiotrophoblasts and significantly increased in the placenta during the development of pre-eclampsia. In contrast, miR-515-5p overexpression inhibited the differentiation of the syncytiotrophoblast [33].
Inno et al., in their 2021 research, assessed the expression profile of the human miRNome and the dynamics of its changes in the placenta of pregnant women in three trimesters, and looked for relationships with the occurrence of pregnancy complications. Among the obtained conclusions, they pointed out that most of the C19MC miRNA target genes were involved in cell signaling or transcription regulation important in early pregnancy. Accordingly, two thirds of the C19MC miRNA is expressed especially in the first trimester, is very low in the second trimester and slowly increases in the third trimester [29]. Additional research suggested that the advantage of expression primate, paternal-specific C19MC in the first trimester was likely to be associated with a dose-dependent effect on placental transcripts [34][35].
Hromadnikova et al., on the basis of their conducted research, held that C19MC miRNAs were expressed only in placental tissues (miR-520a, miR-516-5p, miR-517, miR-518b, miR-519a, miR-524-5p, miR-525, miR- 526a, miR-526b, miR-520h) [36][37][38]. They also demonstrated the importance of C19MC in the development of placenta-related complications (pre-eclampsia) and pregnancy hypertension or fetal growth restriction (IUGR) [37]. In subsequent studies, Hromadnokova et al. indicated a strong correlation between the increased expression in the first trimester of miR-516-5p, miR-517, and especially miR-520h and miR-518b, and the risk of gestational hypertension [39]. The positive correlation between the increase in expression in the first trimester (12–14 hbd) of miR-520a in the serum of pregnant women who developed severe pre-eclampsia was also previously reported by Ura et al. [40]. Further studies by Hromadnikova et al. confirmed the increased expression of miR-517-5p, miR-518b and miR-520h in the serum of pregnant women tested in the first trimester (11–13 hbd) who developed pre-eclampsia. Herein, miR-517-5p had the highest predictive value. Unfortunately, no correlation was found between the level of C19MC expression and the risk of IUGR [12].
Miura et al. assessed the level of expression miR-520a-5p, miR-520h, miR-516a-5p, miR-516b, miR-518b, miR-519d, miR-525-5p, miR-515-5p, miR-526b, miR-1323 in the plasma of pregnant women at 27–34 weeks of gestation. They observed upregulation of expression of C19MC miRNAs in pregnant women with severe pre-eclampsia (sPE) [41]. Jiang et al., in turn, noticed in a patient with sPE, an increased concentration of miR-520g in the serum already in the first trimester [42]. In two studies from 2014 and 2015, an inverse correlation between the expression level of miRNAs on C19MC and the weight of the placenta and birth weight of newborns was found [41][43]. Other researchers have reported that the level of C19MC expression increases with the advancement of pregnancy [36][38][44] and is higher in the case of early onset PE ((PEEO); <34 weeks of gestation) than late onset PE ((PELO); >37 weeks of gestation) [43].
Chaiwangyen et al. indicated the importance of miR-519d-3p in the formation of immune tolerance in pregnancy by influencing the proliferation and migration of maternal immune cells (monocytes, granulocytes, T-lymphocytes and NK cells) [45]. However, it should be emphasized that humoral factors and miRNAs contained in exosomes also affect the maternal vascular endothelium, stimulating it to release cytokines and activating neutrophil adhesion, and as a result, inducing a systemic inflammatory response characteristic of advanced PE [17]. In addition, Delorme Axford et al. clearly indicated that C19MC miRNAs (miR517-3p, miR-512-3p, or 516b-5p) can increase the resistance of maternal cells at the fetal–mother interface to viral infection, by induction of autophagy [46][47]. Moreover, miR-517a-3p was found to influence the activation of maternal T-lymphocyte and NK cell proliferation, and via the PRKG1 gene, the on activation of the nitric oxide/cGMP signaling pathway [47].
Zhao Z. et al., in their review on the use of miRNAs as potential biomarkers for assessing the risk of pregnancy complications, pointed to conflicting data on the expression of individual miRNAs in the various cited studies. As reasons for this, they cited the possible impact of the following heterogeneity factors in patient populations: ethnic origin; variability in the severity of PE; variability of the gestational age; maternal interview; route of delivery or other test conditions: origin and processing samples (tissues, cells, serum and plasma); and data analysis [31]. Furthermore, they pointed out that the use of C19MC as a biomarker of pre-eclampsia development is somewhat complicated by the fact that some miRNAs: miR-16; let-7d; miR-520a *; miR-520h; miR-525; miR-516-5p; miR-517 *; and miR-518b are not stable enough during long-term frozen plasma storage [48].
In another article from 2019, Hromadnikova et al. indicated a higher predictive value of C19MC miRNAs expression assay using maternal serum exosomes, compared to assaying C19miRNAs expression in whole maternal serum samples. The selected miRNAs expressed only in the placenta with the highest predictive value (miR-516b-5p, miR-517-5p, miR-518b, miR-520a-5p, miR -525-5p) were analyzed in the samples from patients during the first trimester of pregnancy. In patients who subsequently developed GH or PE, decreased expression of miR-517-5p, miR-520a-5p and miR-525-5p was observed. Moreover, decreased expression of miR-520a-5p was found to be correlated with FGR. An important observation is the convergence of the results from maternal serum exosomes with the level of miRNA expression in the postpartum placenta [13].
Analysis of the expression level of C19MC miRNAs in placental tissues obtained after delivery also appeared in an earlier original study by Hromadnikova et al., from 2015. The expression of 15 miRNAs was assessed when the research team was attempting to determine correlations with the development of GH, PE, FGF. In the work, correlation was found between the decreased expression of miR-517-5p, miR-519d, miR-520a-5p and miR-525 and the development of GH and between the decreased expression of miR-517-5p, miR-518f-5p, miR-519a, miR-519d, miR-520a-5p and miR-525 and the development of FGF. Accordingly, the development of PE was associated with a decrease in the expression of miR-515-5p, miR-517-5p, miR-518b, miR-518f-5p, miR-519a, miR-519d, miR-520a-5p, miR-520h, miR-524-5p, miR-525 and miR-526a and was more pronounced the longer this complication of pregnancy lasted. Downregulation of miR-519a expression was also found to be strongly associated with development of severe pre-eclampsia (sPE) [49].
In contrast, in a study on the analysis of the expression profile of the placental miRNAome in all three trimesters, Inno et al. observed in pregnancies complicated with PE, an increase in the expression of 13 C19MC miRNAs with a negative correlation with gene expression. The strongest correlation was found for the expression of miR-522-5p and miR-518a-5p [35].
A careful analysis of the role of individual C19MC miRNAs and their target genes may explain at what stage and how they are involved in the development of pre-eclampsia. Buckberry et al. tried to systematize the knowledge about the importance of the variable expression of C19MC miRNAs in the development of pre-eclampsia through the analysis of selected target genes [50]. Their studies consistently showed an increase in expression of eight of the C19MC miRNAs during the development of pre-eclampsia. In addition, it was shown that miR-520g and miR-520h inhibited the expression of VEGF and a simultaneous increase in the expression of the VEGF receptor gene, FLT1, in pre-eclamptic placentas [51]. In turn, the importance of increased expression of selected miRNAs C19MC in pre-eclampsia in terms of apoptosis regulation or modification of pro-apoptotic factors is probably related to the CDKN1A (p21) gene. For miR-519b, 519e, miR-520h and possibly miR-517a, the CDKN1A (p21) gene is a target gene [52]. CDKN1A (p21) is associated with apoptosis and plays a role as the cell cycle inhibitor.
The correlation between the increase in the expression of C19MC miRNAs in the course of pre-eclampsia, regardless of the time of symptom onset and their severity, is also probably related to target genes, most of which are genes involved in the processes of immune regulation or inflammatory response in the human body [53]. When comparing the expression level of C19MC and the level of genes regulated by miRNAs, a negative correlation can be seen. Hromadnikova et al. placed PAPPA among the mentioned target genes, the expression of which is regulated by miR-517 *. In their work, the PAPPA protein was used in the first trimester screening test [39].
In further work, upregulated miR-519d was suggested to supposedly silence the expression of MMP2, CXCL6, NR4A2 and FOXL2, and was found to be involved in cellular migration and invasion [17][54][55]. In addition, miR-520g is thought to partially inhibit MMP2 synthesis [42], which may lead to impaired remodeling of spiral arteries and thus contribute to the occurrence of PE.
The work of Zhang et al. showed that the target genes for miR-515-5p included hCYP19A1/aromatase, transcription factor glial cells lacking 1 and the WNT receptor ‘frizzled 5’. According to the study, the aforementioned factors played important roles in the process of trophoblast differentiation in early pregnancy [33].
Logan et al. pointed out that the Cajal Bodies marker protein (coilin) was a positive regulator of miR-517-3p biogenesis and was induced by hypoxia. Their study suggested that high expression level of miR-517-3p inhibited the translation of TNFAIP3-Interacting Protein 1 (TNIP1), an inhibitor of the NF-kappa B signaling pathway. Moreover, high levels of miR-517-3p and NF-kappa B inhibit trophoblast invasion and increase sFlt-1 secretion at the same time [56].
Liu et al. showed that miR-518b stimulated trophoblast cell proliferation via the Rap1b-Ras-MAPK pathway. What is more, an increase in the level of observed miR-518b in the PE placenta may lead to excessive trophoblast proliferation [57]. In turn, Canfield et al. investigated the importance of RNA-binding protein LIN28B in the development of PE. They found that there was a decrease in the level of LIN28B in the placenta of women with PE, as compared to normal pregnancies, and that the value of LIN28B was lower the longer the complication lasted [58].
In related work, the knockdown of LIN28B in the JEG3 cell line was seen to reduce cell proliferation, suppress the syncytin 1 (SYN-1) involved in syncytiotrophoblast formation and the apelin receptor endogenous ligand (ELABELA), to decrease C19MC miRNA expression (miR516a, miR-516b and miR-519d) and increase mRNA expression ITGb4 and TNF-a, also influencing the process of inflammation in the placenta [58]. It is believed that the effect of LIN28 on the regulation of C19MC miRNAs expression results from direct binding to the consensus DNA sequence in the promoter regions, and from activation of CpG TET1 demethylase, as well as from binding to the CpG-rich Alu repeats distributed in C19MC, which act as independent promoters of RNA polymerase III [58][59][60].
Liu et al., in other work, showed that miR-520c-3p could block inflammasome activation and the development of the inflammatory cascade in pre-eclampsia by downgrading NLRP3 expression [61]. Xie L. and Sadovsky Y. revealed that miR-519d is a factor that regulated the expression of protein genes involved in the interactions of cells with the extracellular matrix and the processes of migration and thus cell invasion, with no effect on proliferation and apoptosis [62]. Of interest in this work is that it demonstrates the importance of the cell migration process in invasion during trophoblast implantation and infiltration, as well as metastasis during neoplasm [12][42]. The importance of selected C19MC miRNAs in the process of trophoblast development and their modulating effect on the maternal immune response are summarized in Figure 1.
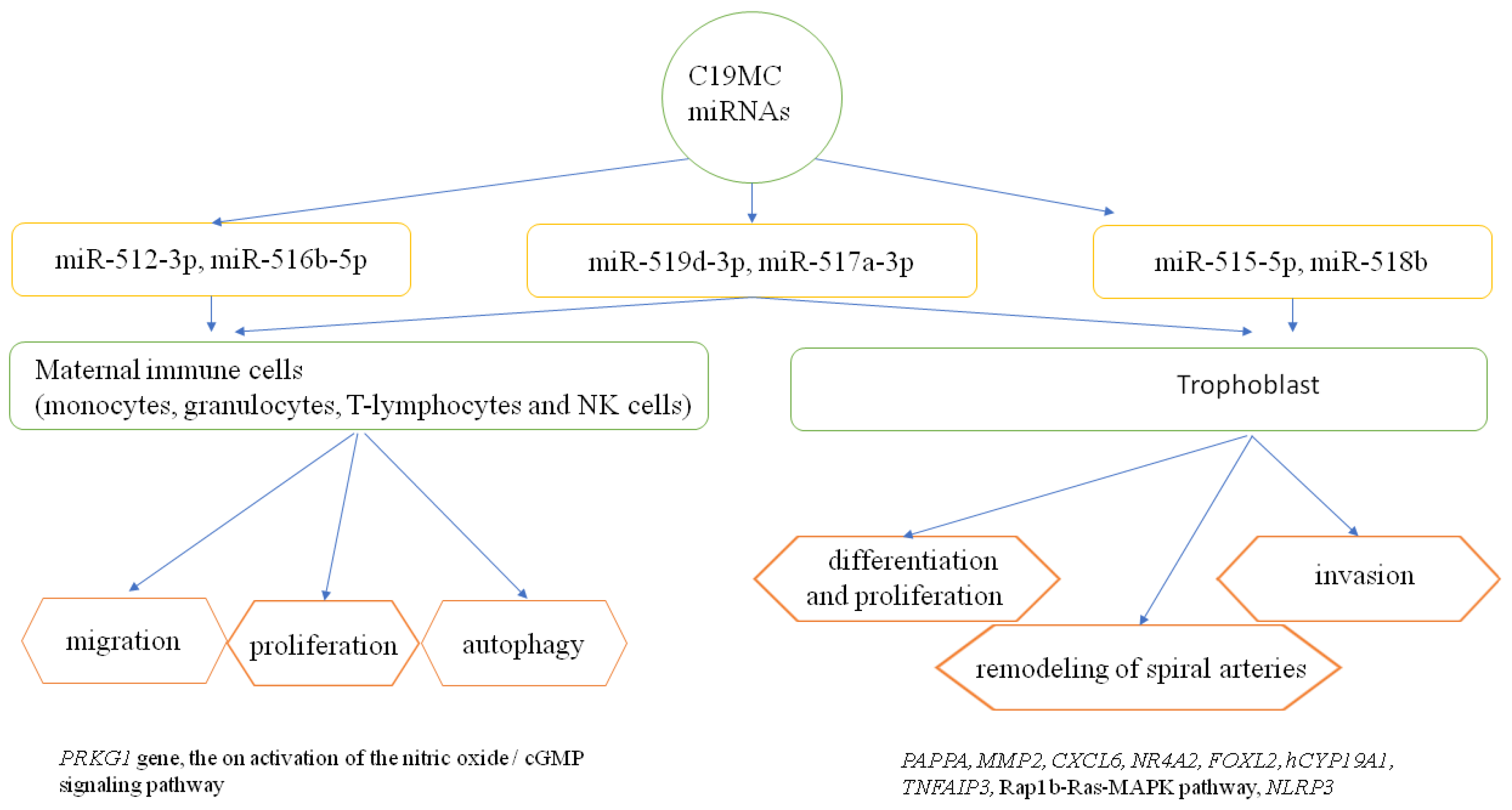
Figure 1. The role of selected C19MC miRNAs in the regulation of trophoblast development and the maternal immune response [12][28][32][33][35][38][39][45][63][64]. PRKG1—protein kinase, cGMP- dependent, regulatory, type 1; PAPPA—pappalysin 1, pregnancy-associated plasma protein A; MMP2—matrix metalloproteinase 2; CXCL6—chemokine, CXC motif, ligand 6; NR4A2—nuclear receptor subfamily 4, group A, member 2; FOXL2—forkhead transcription factor FOXL2; hCYP19A1—human cytochrome P450, family 19, subfamily A, polypeptide 1; TNFAIP3—tumor necrosis factor-alpha-induced protein 3; Rap1b-Ras-MAPK pathway-RAS-related protein Rap1b-Ras-mitogen-activated protein kinases pathway; NLRP3—NLR family, pyrin domain-containing 3.
Data from various studies assessing the expression level of C19MC microRNAs in tumors indicate their role in regulating cell proliferation, angiogenesis and possible oncogenic or suppressor activity [65][66][67]. An important common feature of the early stage of placenta development (5–12 hbd) and tumor biology is intense angiogenesis under conditions of relative hypoxia [68][69].
In some aggressive brain tumors, C19MC amplification has been shown [70], while in some thyroid adenomas, the 19q13 region has been involved in chromosomal translocations [4]. Depending on the type of tumor, miR-519d has either an oncogenic or a tumor suppressive function [71]. Here, oncogenic function is related to miR-519d upregulation and has been demonstrated in hepatocellular carcinoma (HCC) [72][73], cervical cancer [67][74] and multiple myeloma [75]. In contrast, tumor suppression function was confirmed in hepatocellular carcinoma [76], lung adenocarcinoma [77], human osteosarcoma [78], ovarian cancer [79], breast cancer [66][80] and chondrosarcoma [81]. Other studies indicate that the regulatory functions of individual C19MC miRNAs in cancer are related to silencing the expression of factors and signaling pathways related to adhesion, migration, differentiation, growth and angiogenesis (Rap1b, ABCG2, DAPK2, ephrins-EphB2 and EphB4, CXCR4) [65][66][67][82][83][84][85].
References
- Bortolin-Cavaillé, M.-L.; Dance, M.; Weber, M.; Cavaillé, J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009, 37, 3464–3473.
- Malnou, C.; Umlauf, D.; Mouysset, M.; Cavaillé, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2019, 9, 706.
- Vaira, V.; Elli, F.M.; Forno, I.; Guarnieri, V.; Verdelli, C.; Ferrero, S.; Scillitani, A.; Vicentini, L.; Cetani, F.; Mantovani, G.; et al. The microRNA cluster C19MC is deregulated in parathyroid tumours. J. Mol. Endocrinol. 2012, 49, 115–124.
- Rippe, V.; Dittberner, L.; Lorenz, V.N.; Drieschner, N.; Nimzyk, R.; Sendt, W.; Junker, K.; Belge, G.; Bullerdiek, J. The Two Stem Cell MicroRNA Gene Clusters C19MC and miR-371-3 Are Activated by Specific Chromosomal Rearrangements in a Subgroup of Thyroid Adenomas. PLoS ONE 2010, 5, e9485.
- Flor, I.; Bullerdiek, J. The dark side of a success story: microRNAs of the C19MC cluster in human tumours. J. Pathol. 2012, 227, 270–274.
- Augello, C.; Vaira, V.; Caruso, L.; Destro, A.; Maggioni, M.; Park, Y.N.; Montorsi, M.; Santambrogio, R.; Roncalli, M.; Bosari, S. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012, 32, 772–782.
- Mouillet, J.-F.; Chu, T.; Sadovsky, Y. Expression patterns of placental microRNAs. Birth Defects Res. Part A Clin. Mol. Teratol. 2011, 91, 737–743.
- Luo, S.-S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human Villous Trophoblasts Express and Secrete Placenta-Specific MicroRNAs into Maternal Circulation via Exosomes. Biol. Reprod. 2009, 81, 717–729.
- Morales-Prieto, D.M.; Favaro, R.R.; Markert, U.R. Placental miRNAs in feto-maternal communication mediated by extracellular vesicles. Placenta 2020, 102, 27–33.
- Donker, R.B.; Mouillet, J.F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012, 18, 417–424.
- Mouillet, J.-F.; Chu, T.; Hubel, C.A.; Nelson, D.M.; Parks, W.T.; Sadovsky, Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta 2010, 31, 781–784.
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756.
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972.
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750.
- Mittelbrunn, M.; Sánchez-Madrid, F. Intercellular communication: Diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 2012, 13, 328–335.
- Redman, C. The six stages of pre-eclampsia. Pregnancy Hypertens. 2014, 4, 246.
- Munjas, J.; Sopić, M.; Stefanović, A.; Košir, R.; Ninić, A.; Joksić, I.; Antonić, T.; Spasojević-Kalimanovska, V.; Zmrzljak, U.P. Non-Coding RNAs in Preeclampsia—Molecular Mechanisms and Diagnostic Potential. Int. J. Mol. Sci. 2021, 22, 10652.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Žarković, M.; Hufsky, F.; Markert, U.R.; Marz, M. The Role of Non-Coding RNAs in the Human Placenta. Cells 2022, 11, 1588.
- Tannetta, D.; Sargent, I. Placental Disease and the Maternal Syndrome of Preeclampsia: Missing Links? Curr. Hypertens. Rep. 2013, 15, 590–599.
- Pillay, P.; Maharaj, N.; Moodley, J.; Mackraj, I. Placental exosomes and pre-eclampsia: Maternal circulating levels in normal pregnancies and, early and late onset pre-eclamptic pregnancies. Placenta 2016, 46, 18–25.
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefèvre, A.; Coullin, P.; Moore, G.E.; Cavaillé, J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010, 19, 3566–3582.
- Monk, D.; Arnaud, P.; Apostolidou, S.; Hills, F.A.; Kelsey, G.; Stanier, P.; Feil, R.; Moore, G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 6623–6628.
- Wagschal, A.; Feil, R. Genomic imprinting in the placenta. Cytogenet. Genome Res. 2006, 113, 90–98.
- Davies, W.; Isles, A.R.; Humby, T.; Wilkinson, L.S. What Are Imprinted Genes Doing in the Brain? Adv. Exp. Med. Biol. 2008, 626, 62–70.
- Tycko, B. Imprinted genes in placental growth and obstetric disorders. Cytogenet. Genome Res. 2006, 113, 271–278.
- Smith, F.; Garfield, A.; Ward, A. Regulation of growth and metabolism by imprinted genes. Cytogenet. Genome Res. 2006, 113, 279–291.
- Mohammad, F.; Mondal, T.; Kanduri, C. Epigenetics of imprinted long non-coding RNAs. Epigenetics 2009, 4, 277–286.
- Meaburn, E.; Schalkwyk, L.; Mill, J. Allele-specific methylation in the human genome: Implications for genetic studies of complex disease. Epigenetics 2010, 5, 578–582.
- Chim, S.S.C.; Shing, T.K.F.; Hung, E.C.W.; Leung, T.-Y.; Lau, T.-K.; Chiu, R.W.K.; Lo, Y.M.D. Detection and Characterization of Placental MicroRNAs in Maternal Plasma. Clin. Chem. 2008, 54, 482–490.
- Zhao, Z.; Moley, K.H.; Gronowski, A.M. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin. Biochem. 2013, 46, 953–960.
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770.
- Zhang, M.; Muralimanoharan, S.; Wortman, A.C.; Mendelson, C.R. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl. Acad. Sci. USA 2016, 113, E7069–E7076.
- Haig, D.; Mainieri, A. The Evolution of Imprinted microRNAs and Their RNA Targets. Genes 2020, 11, 1038.
- Inno, R.; Kikas, T.; Lillepea, K.; Laan, M. Coordinated Expressional Landscape of the Human Placental miRNome and Transcriptome. Front. Cell Dev. Biol. 2021, 9, 697947.
- Hromadnikova, I. Extracellular Nucleic Acids in Maternal Circulation as Potential Biomarkers for Placental Insufficiency. DNA Cell Biol. 2012, 31, 1221–1232.
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Doucha, J.; Krofta, L. Circulating C19MC MicroRNAs in Preeclampsia, Gestational Hypertension, and Fetal Growth Restriction. Mediat. Inflamm. 2013, 2013, 186041.
- Kotlabova, K.; Doucha, J.; Hromadnikova, I. Placental-specific microRNA in maternal circulation—identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011, 89, 185–191.
- Hromadnikova, I.; Kotlabova, K.; Hympanova, L.; Doucha, J.; Krofta, L. First Trimester Screening of Circulating C19MC microRNAs Can Predict Subsequent Onset of Gestational Hypertension. PLoS ONE 2014, 9, e113735.
- Ura, B.; Feriotto, G.; Monasta, L.; Bilel, S.; Zweyer, M.; Celeghini, C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan. J. Obstet. Gynecol. 2014, 53, 232–234.
- Miura, K.; Morisaki, S.; Abe, S.; Higashijima, A.; Hasegawa, Y.; Miura, S.; Tateishi, S.; Mishima, H.; Yoshiura, K.; Masuzaki, H. Circulating levels of maternal plasma cell-free pregnancy-associated placenta-specific microRNAs are associated with placental weight. Placenta 2014, 35, 848–851.
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75.
- Miura, K.; Higashijima, A.; Murakami, Y.; Tsukamoto, O.; Hasegawa, Y.; Abe, S.; Fuchi, N.; Miura, S.; Kaneuchi, M.; Masuzaki, H. Circulating chromosome 19 miRNA cluster microRNAs in pregnant women with severe pre-eclampsia. J. Obstet. Gynaecol. Res. 2015, 41, 1526–1532.
- Miura, K.; Miura, S.; Yamasaki, K.; Higashijima, A.; Kinoshita, A.; Yoshiura, K.-I.; Masuzaki, H. Identification of Pregnancy-Associated MicroRNAs in Maternal Plasma. Clin. Chem. 2010, 56, 1767–1771.
- Chaiwangyen, W.; Murrieta-Coxca, J.M.; Favaro, R.R.; Photini, S.M.; Gutiérrez-Samudio, R.N.; Schleussner, E.; Markert, U.R.; Morales-Prieto, D.M. MiR-519d-3p in Trophoblastic Cells: Effects, Targets and Transfer to Allogeneic Immune Cells via Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 3458.
- Delorme-Axford, E.; Donker, R.B.; Mouillet, J.-F.; Chu, T.; Bayer, A.; Ouyang, Y.; Wang, T.; Stolz, D.B.; Sarkar, S.N.; Morelli, A.E.; et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12048–12053.
- Kambe, S.; Yoshitake, H.; Yuge, K.; Ishida, Y.; Ali, M.; Takizawa, T.; Kuwata, T.; Ohkuchi, A.; Matsubara, S.; Suzuki, M.; et al. Human Exosomal Placenta-Associated miR-517a-3p Modulates the Expression of PRKG1 mRNA in Jurkat Cells. Biol. Reprod. 2014, 91, 129.
- Hromadnikova, I.; Kotlabova, K.; Doucha, J.; Dlouha, K.; Krofta, L. Absolute and Relative Quantification of Placenta-Specific MicroRNAs in Maternal Circulation with Placental Insufficiency–Related Complications. J. Mol. Diagn. 2012, 14, 160–167.
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Pirkova, P.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Krofta, L. Expression Profile of C19MC microRNAs in Placental Tissue in Pregnancy-Related Complications. DNA Cell Biol. 2015, 34, 437–457.
- Buckberry, S.; Bianco-Miotto, T.; Roberts, C.T. Imprinted and X-linked non-coding RNAs as potential regulators of human placental function. Epigenetics 2014, 9, 81–89.
- Meng, T.; Chen, H.; Sun, M.; Wang, H.; Zhao, G.; Wang, X. Identification of Differential Gene Expression Profiles in Placentas from Preeclamptic Pregnancies Versus Normal Pregnancies by DNA Microarrays. OMICS A J. Integr. Biol. 2012, 16, 301–311.
- Ishibashi, O.; Ohkuchi, A.; Ali, M.; Kurashina, R.; Luo, S.-S.; Ishikawa, T.; Takizawa, T.; Hirashima, C.; Takahashi, K.; Migita, M.; et al. Hydroxysteroid (17-β) Dehydrogenase 1 Is Dysregulated by Mir-210 and Mir-518c That Are Aberrantly Expressed in Preeclamptic Placentas. Hypertension 2012, 59, 265–273.
- Available online: http://mirdb.org/miRDB/ (accessed on 5 November 2022).
- Xie, L.; Mouillet, J.-F.; Chu, T.; Parks, W.T.; Sadovsky, E.; Knöfler, M.; Sadovsky, Y. C19MC MicroRNAs Regulate the Migration of Human Trophoblasts. Endocrinology 2014, 155, 4975–4985.
- Ding, J.; Huang, F.; Wu, G.; Han, T.; Xu, F.; Weng, D.; Wu, C.; Zhang, X.; Yao, Y.; Zhu, X. MiR-519d-3p Suppresses Invasion and Migration of Trophoblast Cells via Targeting MMP-2. PLoS ONE 2015, 10, e0120321.
- Logan, M.K.; Lett, K.E.; McLaurin, D.M.; Hebert, M.D. Coilin as a regulator of NF-kB mediated inflammation in preeclampsia. Biol. Open 2022, 11, bio059326.
- Liu, M.; Wang, Y.; Lu, H.; Wang, H.; Shi, X.; Shao, X.; Li, Y.-X.; Zhao, Y.; Wang, Y.-L. miR-518b Enhances Human Trophoblast Cell Proliferation Through Targeting Rap1b and Activating Ras-MAPK Signal. Front. Endocrinol. 2018, 9, 100.
- Canfield, J.; Arlier, S.; Mong, E.F.; Lockhart, J.; Van Wye, J.; Guzeloglu-Kayisli, O.; Schatz, F.; Magness, R.R.; Lockwood, C.J.; Tsibris, J.C.M.; et al. Decreased LIN28B in preeclampsia impairs human trophoblast differentiation and migration. FASEB J. 2019, 33, 2759–2769.
- Hafner, M.; Max, K.E.; Bandaru, P.; Morozov, P.; Gerstberger, S.; Brown, M.; Molina, H.; Tuschl, T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 2013, 19, 613–626.
- Zeng, Y.; Yao, B.; Shin, J.; Lin, L.; Kim, N.; Song, Q.; Liu, S.; Su, Y.; Guo, J.U.; Huang, L.; et al. Lin28A Binds Active Promoters and Recruits Tet1 to Regulate Gene Expression. Mol. Cell 2015, 61, 153–160.
- Liu, Z.; Zhao, X.; Shan, H.; Gao, H.; Wang, P. microRNA-520c-3p suppresses NLRP3 inflammasome activation and inflammatory cascade in preeclampsia by downregulating NLRP3. Inflamm. Res. 2019, 68, 643–654.
- Xie, L.; Sadovsky, Y. The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation. Placenta 2016, 48, 34–37.
- Morales-Prieto, D.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734.
- Yang, S.; Li, H.; Ge, Q.; Guo, L.; Chen, F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol. Med. Rep. 2015, 12, 527–534.
- Keung, M.-H.; Chan, L.-S.; Kwok, H.-H.; Wong, R.N.-S.; Yue, P.Y.-K. Role of microRNA-520h in 20(R)-ginsenoside-Rg3-mediated angiosuppression. J. Ginseng Res. 2016, 40, 151–159.
- Su, C.-M.; Wang, M.-Y.; Hong, C.-C.; Chen, H.-A.; Su, Y.-H.; Wu, C.-H.; Huang, M.-T.; Chang, Y.-W.; Jiang, S.-S.; Sung, S.-Y.; et al. miR-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene 2016, 35, 1134–1142.
- Chang, Y.-W.; Chen, M.-W.; Chiu, C.-F.; Hong, C.-C.; Cheng, C.-C.; Hsiao, M.; Chen, C.-A.; Wei, L.-H.; Su, J.-L. Arsenic Trioxide Inhibits CXCR4-Mediated Metastasis by Interfering miR-520h/PP2A/NF-κB Signaling in Cervical Cancer. Ann. Surg. Oncol. 2014, 21, 687–695.
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239.
- Jaszczuk, I.; Koczkodaj, D.; Kondracka, A.; Kwaśniewska, A.; Winkler, I.; Filip, A. The role of miRNA-210 in pre-eclampsia development. Ann. Med. 2022, 54, 1350–1356.
- Li, M.; Lee, K.F.; Lu, Y.; Clarke, I.; Shih, D.; Eberhart, C.; Collins, V.P.; Van Meter, T.; Picard, D.; Zhou, L.; et al. Frequent Amplification of a chr19q13.41 MicroRNA Polycistron in Aggressive Primitive Neuroectodermal Brain Tumors. Cancer Cell 2009, 16, 533–546, Erratum in Cancer Cell 2010, 17, 413.
- Flor, I.; Neumann, A.; Freter, C.; Helmke, B.M.; Langenbuch, M.; Rippe, V.; Bullerdiek, J. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem. Biophys. Res. Commun. 2012, 422, 411–416.
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Marasco, E.; Capranico, G.; Mantovani, V.; Marinello, J.; Sabbioni, S.; Callegari, E.; et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J. Pathol. 2012, 227, 275–285.
- Fornari, F.; Ferracin, M.; Trerè, D.; Milazzo, M.; Marinelli, S.; Galassi, M.; Venerandi, L.; Pollutri, D.; Patrizi, C.; Borghi, A.; et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS ONE 2015, 10, e0141448.
- Zhou, J.-Y.; Zheng, S.-R.; Liu, J.; Shi, R.; Yu, H.-L.; Wei, M. MiR-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int. 2016, 16, 21.
- Berenstein, R.; Blau, O.; Nogai, A.; Waechter, M.; Slonova, E.; Schmidt-Hieber, M.; Kunitz, A.; Pezzutto, A.; Doerken, B.; Blau, I.W. Multiple myeloma cells alter the senescence phenotype of bone marrow mesenchymal stromal cells under participation of the DLK1-DIO3 genomic region. BMC Cancer 2015, 15, 68.
- Hou, Y.-Y.; Cao, W.-W.; Li, L.; Li, S.-P.; Liu, T.; Wan, H.-Y.; Liu, M.; Li, X.; Tang, H. MicroRNA-519d targets MKi67 and suppresses cell growth in the hepatocellular carcinoma cell line QGY-7703. Cancer Lett. 2011, 307, 182–190.
- Gennarino, V.A.; D’Angelo, G.; Dharmalingam, G.; Fernandez, S.; Russolillo, G.; Sanges, R.; Mutarelli, M.; Belcastro, V.; Ballabio, A.; Verde, P.; et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012, 22, 1163–1172.
- Tsai, H.-C.; Su, H.-L.; Huang, C.-Y.; Fong, Y.-C.; Hsu, C.-J.; Tang, C.-H. CTGF increases matrix metalloproteinases expression and subsequently promotes tumor metastasis in human osteosarcoma through down-regulating miR-519d. Oncotarget 2014, 5, 3800–3812.
- Pang, Y.; Mao, H.; Shen, L.; Zhao, Z.; Liu, R.; Liu, P. MiR-519d represses ovarian cancer cell proliferation and enhances cisplatin-mediated cytotoxicity in vitro by targeting XIAP. OncoTargets Ther. 2014, 7, 587–597.
- Deng, X.; Zhao, Y.; Wang, B. miR-519d-mediated downregulation of STAT3 suppresses breast cancer progression. Oncol. Rep. 2015, 34, 2188–2194.
- Tsai, C.-H.; Tsai, H.-C.; Huang, H.-N.; Hung, C.-H.; Hsu, C.-J.; Fong, Y.-C.; Hsu, H.-C.; Huang, Y.-L.; Tang, C.-H. Resistin promotes tumor metastasis by down-regulation of miR-519d through the AMPK/p38 signaling pathway in human chondrosarcoma cells. Oncotarget 2015, 6, 258–270.
- Zhang, M.; Zhou, S.; Zhang, L.; Zhang, J.; Cai, H.; Zhu, J.; Huang, C.; Wang, J. miR-518b is down-regulated, and involved in cell proliferation and invasion by targeting Rap1b in esophageal squamous cell carcinoma. FEBS Lett. 2012, 586, 3508–3521.
- Frische, E.; Zwartkruis, F. Rap1, a mercenary among the Ras-like GTPases. Dev. Biol. 2010, 340, 1–9.
- Ribeiro-Neto, F.; Urbani, J.; Lemee, N.; Lou, L.; Altschuler, D.L. On the mitogenic properties of Rap1b: cAMP-induced G1/S entry requires activated and phosphorylated Rap1b. Proc. Natl. Acad. Sci. USA 2002, 99, 5418–5423.
- Wang, F.; Xue, X.; Wei, J.; An, Y.; Yao, J.; Cai, H.; Wu, J.; Dai, C.; Qian, Z.; Xu, Z.; et al. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br. J. Cancer 2010, 103, 567–574.
More
Information
Subjects:
Obstetrics & Gynaecology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
914
Revisions:
2 times
(View History)
Update Date:
30 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




