Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kenneth Nugent | -- | 1684 | 2022-11-27 20:12:00 | | | |
| 2 | Conner Chen | Meta information modification | 1684 | 2022-11-30 08:46:26 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Songtanin, B.; Peterson, C.J.; Molehin, A.J.; Nugent, K. Biofilm Formation Based on Studies with Pseudomonas aeruginosa. Encyclopedia. Available online: https://encyclopedia.pub/entry/36675 (accessed on 07 February 2026).
Songtanin B, Peterson CJ, Molehin AJ, Nugent K. Biofilm Formation Based on Studies with Pseudomonas aeruginosa. Encyclopedia. Available at: https://encyclopedia.pub/entry/36675. Accessed February 07, 2026.
Songtanin, Busara, Christopher J. Peterson, Adebayo J. Molehin, Kenneth Nugent. "Biofilm Formation Based on Studies with Pseudomonas aeruginosa" Encyclopedia, https://encyclopedia.pub/entry/36675 (accessed February 07, 2026).
Songtanin, B., Peterson, C.J., Molehin, A.J., & Nugent, K. (2022, November 27). Biofilm Formation Based on Studies with Pseudomonas aeruginosa. In Encyclopedia. https://encyclopedia.pub/entry/36675
Songtanin, Busara, et al. "Biofilm Formation Based on Studies with Pseudomonas aeruginosa." Encyclopedia. Web. 27 November, 2022.
Copy Citation
Studies with the bacterium P. aeruginosa provide important information about the attachment process. One important consideration is how bacteria sense a surface. In P. aeruginosa, there are two distinct surface sensing mechanisms. The first one involves a WSP chemosensory system. In response to the surface contact, the WSP system stimulates the production of cyclic diguanylate. The initial event is uncertain; it probably involves distortion of the cell membrane, which then activates membrane proteins. A second surface sensing mechanism involves type 4 pili. Upon contact with the surface, the methyl-accepting chemotaxis protein Pil J transduces a signal to the protein CyaB, stimulating its activity.
biofilm
microbiome
interstitial epithelial cells
1. Introduction
Biofilms make important contributions to the environment, industry, and health [1][2]. The composition of biofilms includes bacteria adherent to surfaces embedded in an extracellular matrix composed of polysaccharides, proteins, nucleic acids, and nutrients [3]. Biofilms frequently develop on devices, such as prostheses and intravascular catheters, used during patient care. These can result in infections that are difficult to manage due to increased resistance to routine antibiotics. Studies of biofilms on human surfaces are hampered by the need for simple and safe access to the surfaces. The best study location involves the development of biofilms on oral structures that can lead to gingivitis, dental plaque, and dental caries.
2. Biofilm Formation Based on Studies with Pseudomonas aeruginosa
2.1. Biofilm Structure
The initial step in biofilm formation involves the reversible attachment of free-floating bacteria (planktonic) or other microbes to an abiotic or biotic surface (Figure 1) [3][4]. The initial attachment may involve hydrophobic forces. This initial step or stage 1 is reversible, and the attachment may not persist. However, some bacteria undergo a phenotypic change and subsequently adhere to the surface (stage 2). This can result in a formation of more flagella and pili, which create more attachment structures. These bacteria replicate, and small microcolonies are formed (stage 3). The bacteria form an extracellular matrix network that consists of proteins, polysaccharides, DNA, and exogenous debris in the region of attachment. These microcolonies then grow into a biofilm with increased numbers of bacteria and extracellular matrix (stage 4).
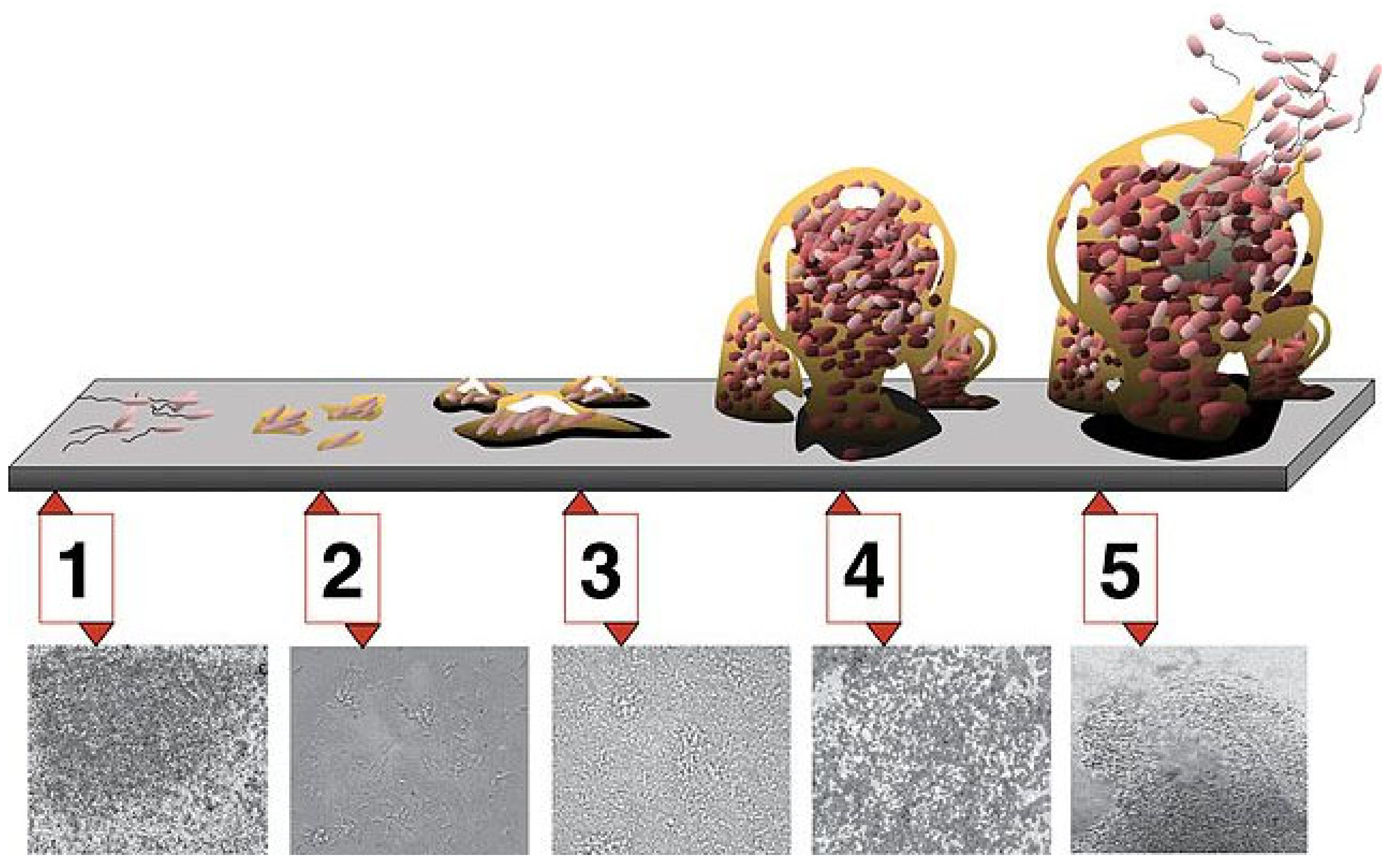
Figure 1. Stages in the development of a biofilm. Stage 1 involves adherence, stage 2 involves attachment and replication, stage 3 involves microcolony formation, stage 4 involves the production of the matrix with biofilm formation, and stage 5 involves partial dispersion of the biofilm, with the free-floating bacteria potentially forming new biofilms [5].
Approximately 85% of the biofilm constitutes the extracellular matrix. The biofilm includes live bacteria, dead bacteria, and relatively quiescent bacteria or persisters. In addition, some biofilms contain multiple bacterial species, including bacteria that cannot independently form biofilms. Eventually, enzymes are released that break down part of the matrix and allow the dispersion of bacteria back into a free-floating (planktonic) existence (stage 5). These bacteria can then attach to other surfaces and start the formation of new biofilms.
Kragh studied biofilm formation using either single bacterial cells or aggregates as the initial source of the biofilms using in vitro culture methods and confocal microscopy to monitor growth [6]. With increasing cell density, aggregates grow better than single cells. Growth rates in aggregates depend on the location of the bacteria in the aggregate, and bacteria in the center of the aggregate have fewer progeny. Bacteria on the surface of the aggregate have much faster and better growth rates. Their study demonstrates that biofilm formation is enhanced when the biofilm is initially seeded by aggregates rather than single cells. In addition, growth in an aggregate depends on cellular location, which presumably affects access to nutrients. Paula et al. analyzed the dynamics of bacterial population growth in biofilms using in vitro growth techniques and confocal microscopy [7]. They noted that bacteria settle randomly on surfaces, but only a subset of these bacteria grows. These active bacteria grow in a three-dimensional pattern by incorporating nearby bacteria into densely populated microcolonies. This clustering and microcolony formation depend on exopolysaccharides to provide a supporting structure. In addition, growth depends on environmental conditions and the availability of nutrients.
Reichhardt used confocal laser scanning microscopy to analyze P. aeruginosa biofilm architecture and matrix localization [8]. The use of this microscopy can help investigators determine the structure of biofilms and the localization of certain components of the biofilm. Pseudomonas biofilms contain high levels of Psl polysaccharides, Pel polysaccharides, alginate, and extracellular DNA. During the formation of biofilms, some of these structural elements change location. For example, Psl is initially located near the bacteria forming the initial matrix. However, as the biofilm grows, Psl moves toward the periphery. Nutrients must diffuse into the biofilm matrix, and this process depends on the diffusion coefficient of the molecule and the dimensions of the biofilm matrix [9]. In addition, some bacteria form microchannels in the biofilm which allows the diffusion of molecules [10]. Studies with antibiotics provide another method of monitoring molecular diffusion into the matrix and are relevant to understanding that antibiotic resistance is associated with biofilms. For example, ciprofloxacin rapidly penetrates into biofilms during static incubation. However, tobramycin accumulates on the periphery of the biofilm aggregate and is tightly adherent to various structures since it is not removed by rinsing the biofilm.
2.2. Bacterial Attachment to Surfaces
Studies with the bacterium P. aeruginosa provide important information about the attachment process (Figure 2). One important consideration is how bacteria sense a surface [11]. Following irreversible attachment, cells start to multiply and produce the matrix. In Gram-negative bacteria, the key intracellular signaling molecule is cyclic diguanylate. With high levels of cyclic diguanylate, bacteria produce more of a biofilm matrix and have less flagella-mediated swimming motility. In P. aeruginosa, there are two distinct surface sensing mechanisms [12]. The first one involves a WSP chemosensory system [13]. In response to the surface contact, the WSP system stimulates the production of cyclic diguanylate. The initial event is uncertain; it probably involves distortion of the cell membrane, which then activates membrane proteins. A second surface sensing mechanism involves type 4 pili. Upon contact with the surface, the methyl-accepting chemotaxis protein Pil J transduces a signal to the protein CyaB, stimulating its activity [14]. This protein increases cellular levels of cyclic adenosine monophosphate (AMP), which increases type 4 pili production and twitching motility. PilY1 has a von Willebrand motif that might have a mechanosensory function [15]. However, in both systems, it is unclear what the exact signal is that is triggered by contact. It is thought that, through successive surface interaction or attachment and detachment, cells become surface adapted [16]. This results in a progressive increase in cellular cyclic AMP and a gradual increase in type 4 pili.
Pili support several essential physiological processes in the gut microbes [17][18][19]. Type 4 pili are involved in the attachment of bacteria to other cells, such as bacteria and epithelial cells, as well as to food and fiber in the gut lumen. Attachment to host cells influences signaling and gene expression and increases the production of cyclic AMP and cyclic diguanylate. Pili are involved in adhesion, biofilm formation, motility, and molecule exchange, including DNA uptake and protein secretions. These activities increase the residence time of bacteria in the biofilm, allow nutrient exchange, promote horizontal gene transfer, increase survival, and provide protection from physical stressors. DNA exchange between bacteria can occur through three processes, including conjugation, transduction, and natural transformation. This transfer of nucleic acids is mediated by type 4 pili and other filamentous structures. Type 4 pili also allow the secretion of proteins that are essential for the bacterial colony.
Type 4 pili are extracellular filaments 6–8 nm wide and several micrometers long, composed of Pil A subunits that are assembled in a helical manner [12]. The Pil A leader sequence is approximately 10 amino acids; the mature Pil A is 140 to 160 amino acids. After the peptide sequence is oriented correctly in the inner membrane, the leader sequence is cleaved off, and the Pil A peptide is then incorporated into the growing pilus structure. One ATPase attaches the Pil A to the growing pilus; a second ATPase detaches the Pil A from the pilus as it is retracted. Chang and colleagues published a detailed study on the formation and architecture of type 4 pili [20].
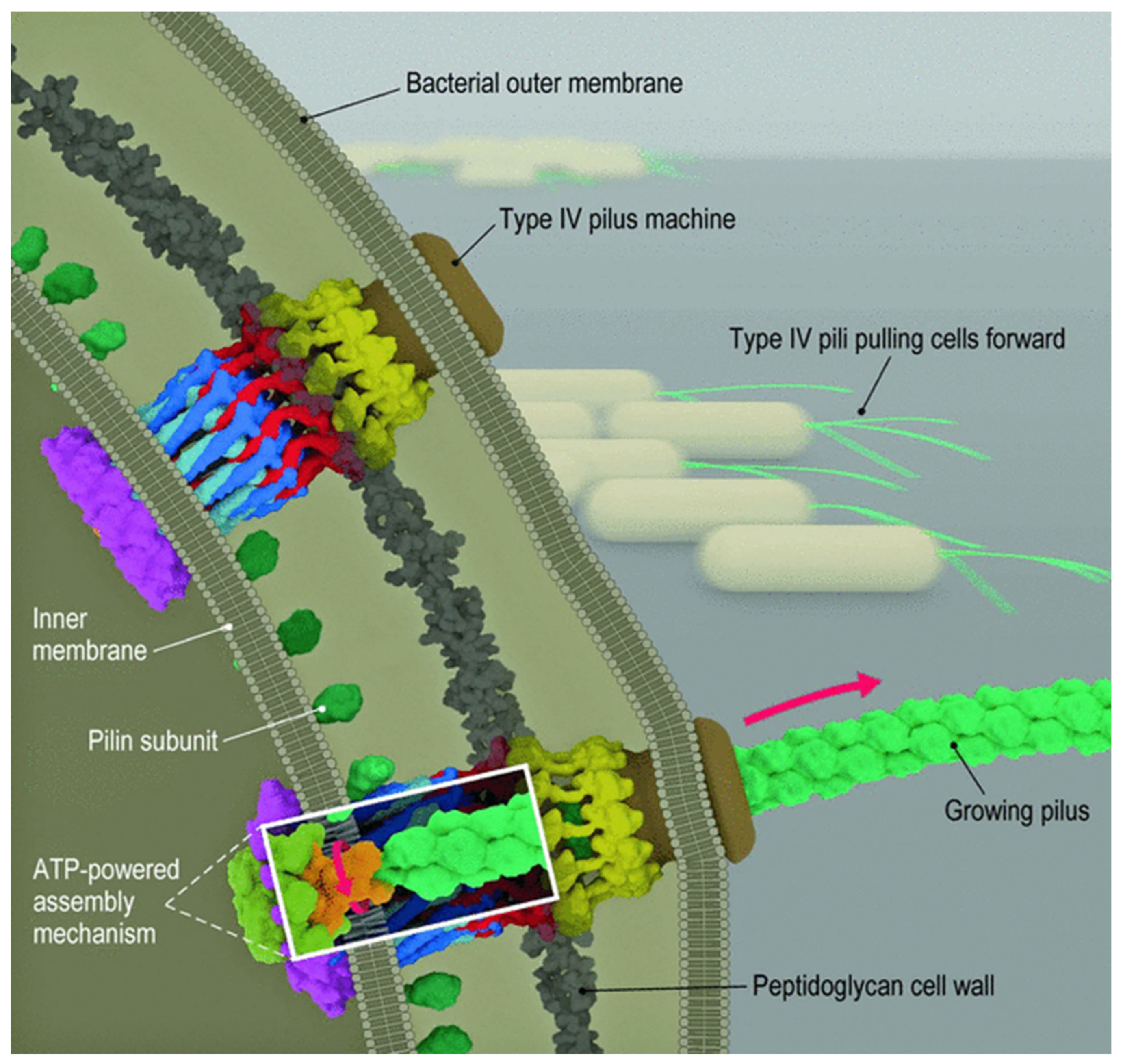
Figure 2. Structure and function of type 4 pili. The addition of peptides to the growing pilus requires ATP for energy. After attachment of the pilus to a surface, retraction of the pilus requires energy provided by ATP. This results in the movement of the bacterium [20].
Retraction of the pilus generates mechanical force and results in motility. Beaussart et al. studied adhesion and mechanical stress of type 4 pili of P. aeruginosa using atomic force microscopy [21]. They demonstrated that these bacteria adhered firmly to hydrophobic surfaces in a time-dependent manner but only weakly adhered to hydrophilic surfaces. PilY1 is a possible pilus-associated adhesin required for colonization of epithelial cells. In their study, the pili had an average length of 1.1 ± 0.3 µm and an average diameter of 4.2 ± 1.1 nm. Multiple force–distance curves demonstrated that these pili had an average adhesion force of 50–250 pN and an average rupture length of 50 to 2000 nm. These stretching experiments resulted in the development of a constant-force region that likely represents uncoiling or unfolding of their helical quaternary structure. Other studies suggested that they can behave as nanosprings. Increasing the adhesion time increases the mean adhesion force to 3000 pN. Studies have also suggested that Pil Y1 may have no direct role in these measurements. These investigators concluded that the adhesion of Pseudomonas to hydrophobic surfaces involves both strong cohesive interactions based on time-dependent interactions over short distances with cell surface constituents, such as membrane proteins and other surface structures, and weaker interactions involving extension and force-induced configuration changes in the type 4 pili over longer distances [21]. Pseudomonas type 4 pili also bind to living pneumocytes [21].
In summary, biofilm formation requires the adherence of bacteria to surface structures, the production of an extracellular matrix, growth of bacteria, and communication between bacteria through the production of quorum sensing of molecules. Metabolic activities in the biofilm include alterations in gene transcription, the uptake of exogenous DNA resulting in new biosynthetic activity and possibly antibiotic resistance, and protein synthesis and secretion. The structure of the biofilm provides a “protective shelter” which increases antibiotic resistance and reduces the interaction of phagocytes with the bacteria. Studies using Pseudomonas spp. have made significant contributions to the understanding of biofilm formation.
References
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108.
- Gadkari, J.; Bhattacharya, S.; Shrivastav, A. Importance and application of biofilm in microbe assisted bioremediation. In Development in Waste Water Treatment Research and Processes; Shah, M.P., Rodriguez-Couto, S., Riti Thapar, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 154–173.
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019, 27, 17–25.
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Biofilms. Int. J. Mol. Sci. 2020, 21, 8671.
- Monroe, D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007, 5, e307.
- Kragh, K.N.; Hutchison, J.B.; Melaugh, G.; Rodesney, C.; Roberts, A.E.; Irie, Y.; Jensen, P.; Diggle, S.P.; Allen, R.J.; Gordon, V.; et al. Role of Multicellular Aggregates in Biofilm Formation. mBio 2016, 7, e00237.
- Paula, A.J.; Hwang, G.; Koo, H. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat. Commun. 2020, 11, 1354.
- Reichhardt, C.; Parsek, M.R. Confocal Laser Scanning Microscopy for Analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 2019, 10, 677.
- Stewart, P.S. Diffusion in biofilms. J. Bacteriol. 2003, 185, 1485–1491.
- Rooney, L.M.; Amos, W.B.; Hoskisson, P.A.; McConnell, G. Intra-colony channels in E. coli function as a nutrient uptake system. ISME J. 2020, 14, 2461–2473.
- Chang, C.Y. Surface Sensing for Biofilm Formation in. Front. Microbiol. 2017, 8, 2671.
- Ligthart, K.; Belzer, C.; de Vos, W.M.; Tytgat, H.L.P. Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili. Trends Microbiol. 2020, 28, 340–348.
- O’Neal, L.; Baraquet, C.; Suo, Z.; Dreifus, J.E.; Peng, Y.; Raivio, T.L.; Wozniak, D.J.; Harwood, C.S.; Parsek, M.R. The Wsp system of Pseudomonas aeruginosa links surface sensing and cell envelope stress. Proc. Natl. Acad. Sci. USA 2022, 119, e2117633119.
- Nolan, L.M.; Cavaliere, R.; Turnbull, L.; Whitchurch, C.B. Extracellular ATP inhibits twitching motility-mediated biofilm expansion by Pseudomonas aeruginosa. BMC Microbiol. 2015, 15, 55.
- Webster, S.S.; Wong, G.C.L.; O’Toole, G.A. The Power of Touch: Type 4 Pili, the von Willebrand A Domain, and Surface Sensing by Pseudomonas aeruginosa. J. Bacteriol. 2022, 204, e0008422.
- Armbruster, C.R.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319.
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836.
- Lee, J.Y.; Tsolis, R.M.; Bäumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, eabp9960.
- Chew, S.-S.; Tan, L.T.-H.; Law, J.W.-F.; Pusparajah, P.; Goh, B.-H.; Ab Mutalib, N.S.; Lee, L.-H. Targeting Gut Microbial Biofilms—A Key to Hinder Colon Carcinogenesis? Cancers 2020, 12, 2272.
- Chang, Y.W.; Rettberg, L.A.; Treuner-Lange, A.; Iwasa, J.; Søgaard-Andersen, L.; Jensen, G.J. Architecture of the type IVa pilus machine. Science 2016, 351, aad2001.
- Beaussart, A.; Baker, A.E.; Kuchma, S.L.; El-Kirat-Chatel, S.; O’Toole, G.A.; Dufrêne, Y.F. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili. ACS Nano 2014, 8, 10723–10733.
More
Information
Subjects:
Microbiology; Infectious Diseases
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
30 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




