Video Upload Options
Osteoporosis is a complex multifactorial condition of the musculoskeletal system. Osteoporosis and osteoporotic vertebral fracture (OVF) are associated with high medical costs and can lead to poor quality of life. Genetic factors are important in determining bone mass and structure, as well as any predisposition for bone degradation and OVF. However, genetic factors are not enough to explain osteoporosis development and OVF occurrence. Epigenetics describes a mechanism for controlling gene expression and cellular processes without altering DNA sequences. The main mechanisms in epigenetics are DNA methylation, histone modifications, and non-coding RNAs (ncRNAs). Recently, alterations in epigenetic mechanisms and their activity have been associated with osteoporosis and OVF.
1. Introduction
Osteoporosis and osteoporotic fractures are associated with high medical costs and can lead to poor quality of life. Osteoporotic fractures can happen from minor trauma, such as slipping or falling. Worldwide, osteoporotic vertebral fracture (OVF) is the most frequent type of osteoporotic fracture [1][2][3][4]. OVF increases in incidence with age and women and is associated with an increased risk of death [2]. Osteoporosis and OVF are influenced by a multifactorial environment, including genetic factors [5][6][7]. Osteoporosis is generally thought to be caused by a reduction in the number of osteoblasts along with preferential differentiation to fat cells in the aged skeleton [3][4]. This may decrease the quantity and functionality of osteoblasts and increase bone marrow fat in aged bones [3][8]. This can result in a reduction in bone formation and bone microarchitecture, leading to additional vertebral fractures and interfering with bone healing and remodeling after fracture [3][9].
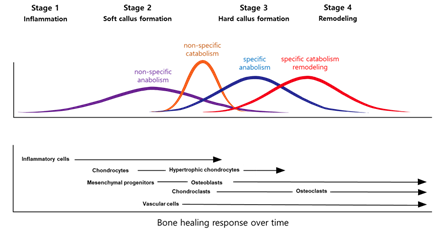
Fracture healing reproduces skeletal development and growth with intricate interactions between cells, growth factors, and extracellular matrices. There are usually four stages to fracture repair: (1) inflammation, (2) soft callus formation, (3) hard callus formation, and (4) remodeling (Figure 1) [10]. Each stage is associated with specific cellular and molecular events [11][12][13]. However, in practice, the events are insufficiently described, and there can be extensive redundancies across the different stages. Further, studies over many years have examined the molecular and cellular forces driving the central process [14][15][16]. When considering cellular processes, vascular cells, inflammatory cells, osteochondral precursors, and osteoclasts maintain an important role in the bone repair process. When considering molecular processes, fracture repair is helped by inflammation-promoting cytokines and three main factors: osteogenesis-promoting factor, growth factor, and angiogenesis factor [17]. These factors help set up relevant morphogenetic disciplines by promoting growth or differentiation and recruiting cells. After that, the damaged soft tissue undergoes a repair and the fracture is covered by soft callus and, subsequently, hard callus.
Figure 1. Models of the fracture healing process.
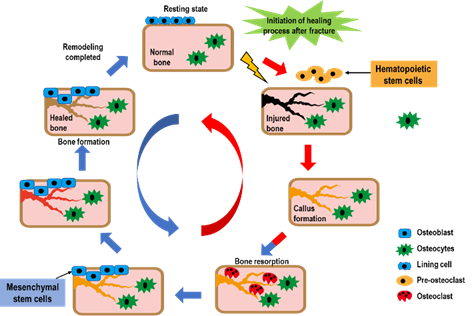
In the final stage of the fracture healing process, remodeling the bone-hardening callus into original cerebellar and cortical structures occurs, and is also known as secondary osteoplasty [13]. This bone remodeling process destroys mineralized bone followed by bone matrix formation that then becomes mineralized. This resorption and formation of bone are highly connected, allowing for skeletal integrity. However, with some pathological conditions such as osteoporosis, bone remodeling steps can uncouple, which can lead to increased fragility and decreased bone strength [18]. To sustain homeostasis of the bone mass, the remodeling of bone integrates extremely regulated steps that rely on the roles and interactions of the osteoblastic and osteoclastic lineages (Figure 2).
Figure 2. The bone healing process after a fracture. Bone remodeling after fracture is initiated by osteoclast activation, and it solubilizes the bone mineral and degrades the matrix. Osteoclasts originate from hematopoietic stem cells which fuse to form multinucleated cells (activated form of osteoclasts). Monocytes/macrophages remove debris, followed by a bone formation phase performed by osteoblasts, producing osteoid matrix which will mineralize.
Bone cell activation and differentiation are regulated at the molecular level by an intricate signaling network that contains systemic hormones and local secreted molecules. It is well known that many probabilistic and environmental stresses can be modulated through phylogenetic determination and gene expression [19][20][21]. Pathological processes for osteopathy with many factors, such as osteopenia or osteoporosis, can have crucial epigenetic elements. Epigenetic elements can present promising areas of research linking disease risk to genetics and gene expression. New treatment for OVF can be developed in virtue of a greater understanding of epigenetic and molecular regulation of bone cell function and differentiation.
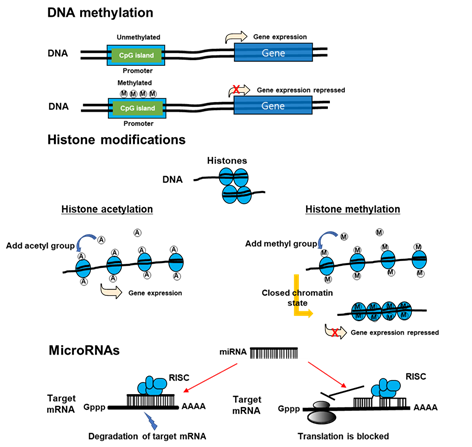
Recently, a new paradigm of epigenetics has advanced. New discipline attempts to identify the delivery of stable phenotypes without altering DNA sequences due to interactions between the genome and internal and external environments. Within epigenetic mechanisms, those that are notable are modifications of histones, DNA methylation, and non-coding RNAs (Figure 3) [22]. Epigenetics can control transcription activity without altering DNA sequences [23] through an enzymatic alteration of 5-cytosine in DNA [24], microRNA [25], histone modifications [26], and remodeling of chromatins. DNA methyltransferases (DNMT) can catalyze methylation of CpG islands of DNA, which can weaken genome stability and reduce the activity of gene transcription [27]. MicroRNA (miRNA) can interrupt mRNA targets and reduce the translation of protein. In addition, it can initiate a post-transcriptional signaling pathway and promote the expression of protein [28][29]. Histone acetyltransferases (HATs) can control histone acetylation, which is necessary to maintain transcription; however, histone deacetylases (HDACs) can eliminate acetyl group from histones, which can favor the formation of heterochromatin to repress the activity of promoters [30].
Figure 3. The mechanisms of epigenetics. DNA methylation inhibits DNA transcription. Histone modification, which modifies the interaction of DNA with histones or other DNA-binding protein complexes, regulates gene expression in a more dynamic and changing manner than does DNA methylation. Non-coding RNAs may be large (>200 nucleotides) or small (18–200 nucleotides). mRNA is one of the small non-coding RNAs. miRNAs suppress gene expression by selectively binding to the 3′ non-coding region (3′UTR) of their mRNA targets through base-paring. miRNAs can negatively regulate gene expression by two different post-transcriptional mechanisms. RISC: RNA induced silencing complex.
Histone methyltransferases catalyze the methylation of histone lysine residues to repress transcription of the gene [31]. Histone demethylases can eliminate the methyl group from lysine, which reverses the transcription of the gene [32]. Beyond enzymatic modification, some metabolites, including succinate, butyrate, and propionate, can trigger histone butyrylation, propionylation, succinylation, and crotonylation [33]. A collective review is necessary of the medical role of mRNA, methylated DNA, and histone modification for bone metabolism and osteoporosis occurrence. This article highlights how DNA methylation, histone modification, and non-coding RNA affect osteoporosis, bone remodeling, and treatment of OVF.
2. Noncoding RNA
Noncoding RNA research, particularly in the musculoskeletal area, is still an unexploited field with a new class being updated regularly. Within the non-coding RNA area, microRNA (miRNA) is the most studied category. miRNA is a small non-coding RNA molecule (containing about 22 nucleotides) found in plants, animals, and some viruses, that modulates gene expression. RNA interference is a powerful mechanism for gene silencing, underlying many aspects of eukaryotic biology. At the molecular level, RNA interference is mediated by a ribonucleoprotein complex called RNA-induced silencing complex (RISC) and can be programmed to target virtually all nucleic acid sequences for silencing. RISC’s ability to find target RNA has been adopted many times in evolution, producing a wide range of gene silencing pathways [34]. Several new functions of miRNAs have been proposed, and several novel classes of larger noncoding RNA with a crucial transcriptional role have been recently demonstrated, including small interfering RNA (siRNA), Piwi-interacting RNAs (piRNAs), long non-coding RNAs (lncRNA), and others (Table 1) [35].
Table 1. The various non-coding RNAs (ncRNA) and their characteristics.
|
ncRNA |
Length (nt) |
Characteristics |
|
MicroRNA(miRNA) |
20–24 |
- Single-stranded RNA derived from pre-miRNA - Silencing of genes |
|
Small interfering RNA (siRNA) |
20–24 |
- Double-stranded RNA processed by endoribonuclease Dicer into mature siRNA - Protection against viral infection - Post-transcriptional silencing/RNA interference |
|
Piwi-interacting RNA (piRNA) |
24–31 |
- Make the complexes with P-element induce wimpy testis (Piwi) proteins of the Argonaute family - Silencing of transposable elements |
|
Promoter-associated RNA (PAR) |
16–200 |
- Single-stranded RNA with half-life - Regulation of post-transcription |
|
Enhancer RNA (eRNA) |
100–9000 |
- Single-stranded RNA with half-life - Activation of transcriptional genes |
|
Long non-coding RNA (lncRNA) |
>200 |
- Non-protein coding transcripts - Modifications of post-transcription - Transcriptional/post-transcriptional regulation - Precursor of siRNA |
2.1. MicroRNAs (miRNAs)
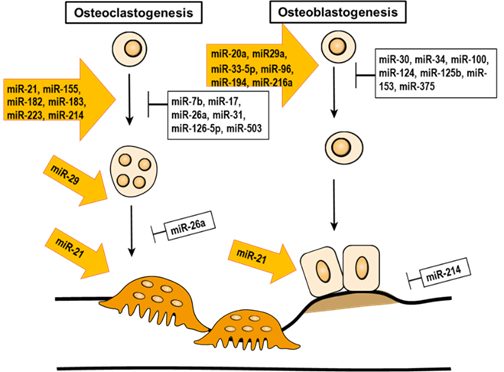
miRNAs are small, non-coding RNA molecules. The size of mature miRNAs ranges between 21–23 nucleotides. Their role is to modulate gene expression by post-transcriptional silencing, either by moderating the cleavage of a targeted transcript or by translational inhibition. In addition, miRNA is involved in the epigenetic regulation of bone remodeling (Table 2 and Figure 3).
Table 2. The pathway/affected molecule of miRNAs and their effects on bone biology.
|
miRNA Family |
Pathway or Affected Molecule |
Effect on Osteoblasts and Osteoclasts |
|
miR-33-5p |
Hmga2 |
Promote the differentiation of osteoblasts |
|
miR-96 |
EGFR, HB-EGF Wnt/β-catenin signaling pathway |
Promote the differentiation of osteoblasts |
|
miR-139-5p |
NOTCH1,Wnt/β-catenin pathway |
Promote the differentiation of osteoblasts |
|
miR-194 |
RUNX2 |
Promote the differentiation of osteoblasts |
|
miR-216a |
PI3K/AKT pathway BMP/TGF-β signaling pathway |
Promote the differentiation of osteoblasts |
|
miRNA-433-3p |
DKK1 |
Promote the differentiation of osteoblasts |
|
miR-542-3p |
SFRP1 BMP-7/PI3K- surviving pathway NKIRAS2, NF-κB signaling pathway |
Promote the differentiation of osteoblasts Inhibit the differentiation of osteoblasts |
|
miR-26a |
Smad1 |
Inhibit the differentiation of osteoblasts |
|
miR-100 |
Smad1 |
Inhibit the differentiation of osteoblasts |
|
miR-124 |
Dlx3, Dlx5, and Dlx2 GSK-3β, Wnt/β-catenin pathway |
Inhibit the differentiation of osteoblasts |
|
miR-125b |
BMPR1b |
Inhibit the differentiation of osteoblasts |
|
miR-153 |
BMPR2 |
Inhibit the differentiation of osteoblasts |
|
miR-203a-3p |
Smad9, Wnt/β-catenin signaling pathway |
Inhibit the differentiation of osteoblasts |
|
miR-214-3p |
ATF4 |
Inhibit the differentiation of osteoblasts |
|
miR-375 |
RUNX2 |
Inhibit the differentiation of osteoblasts |
|
miR-19a |
TWIST and RUNX2 |
Promote the differentiation of osteoclasts |
|
miR-21 |
RANKL,PI3K/Akt signaling pathway, PDCD4 |
Promote the differentiation of osteoclasts |
|
miR-155 |
TNF-α, IL-1β, RANKL, M-CSF, RANK, TRAP, Bcl-2, LEPR, AMPK, p-AMPK, OPG, Bax, TAB 1 |
Promote the differentiation of osteoclasts |
|
miR-182 |
Foxo3, Maml1 |
Promote the differentiation of osteoclasts |
|
miR-183 |
RANKL, HO-1 |
Promote the differentiation of osteoclasts |
|
miR-223 |
TWIST and RUNX2 |
Promote the differentiation of osteoclasts |
|
miR-214 |
Pten, PI3K/Akt pathway |
Promote the differentiation of osteoclasts |
|
miR-7b |
DC-STAMP |
Inhibit the differentiation of osteoclasts |
|
miR-17 |
RANKL |
Inhibit the differentiation of osteoclasts |
|
miR-26a |
CTGF/CCN2 |
Inhibit the differentiation of osteoclasts |
|
miR-31 |
RhoA |
Inhibit the differentiation of osteoclasts |
|
miR-126-5p |
PTHrP and MMP-13 |
Inhibit the differentiation of osteoclasts |
|
miR-141 |
Calcr, EphA2 |
Inhibit the differentiation of osteoclasts |
|
miR-503 |
RANK |
Inhibit the differentiation of osteoclasts |
Hmga2 (high-mobility group AT-hook 2), EGFR (epidermal growth factor receptor), HB-EGF (heparin-binding EGF-like growth factor), NOTCH1 (notch homolog 1, translocation-associated (Drosophila)), RUNX (runt-related transcription factor), PI3K (phosphatidylinositol 3-kinase), AKT (protein kinase B), BMP (bone morphogenetic protein), TGF-β (transforming growth factor beta), DKK1 (Dickkopf Wnt signaling pathway inhibitor 1), SFRP1 (secreted frizzled-related protein 1), NKIRAS2 (NFKB Inhibitor Interacting Ras Like 2), Dlx (Distal-Less Homeobox), GSK-3β (Glycogen synthase kinase 3β), BMPR (bone morphogenetic protein receptor), ATF4 (activating transcription factor 4), TWIST (Twist-related protein), RANKL (receptor activator of nuclear factor kappa-Β ligand), PDCD4 (Programmed cell death protein 4), TNF (tumor necrosis factor), IL (interleukin), M-CSF (macrophage colony-stimulating factor), TRAP (tartrate-resistant acid phosphatase), Bcl-2 (B-cell lymphoma), LEPR (leptin receptor), AMPK (5' adenosine monophosphate-activated protein kinase), OPG (Osteoprotegerin), Bax (Bcl-2-associated X protein), TAB 1 (TGF-beta activated kinase 1 binding protein 1), Foxo3 (forkhead box O3), Maml1 (mastermind-like protein 1), HO-1 (heme oxygenase-1), Pten (phosphatase and tensin homolog), DC-STAMP (dendritic cell-specific transmembrane protein), CTGF/CCN (connective tissue growth factor), RhoA (ras homolog gene family, member A), PTHrP (parathyroid hormone-related peptide), MMP-13 (matrix metallopeptidase 13), Calcr (calcitonin receptor), EphA2 (ephrin type-A receptor 2).
Figure 4. The epigenetic regulation of miRNA in bone remodeling.
RUNX2 is a central transcription factor that regulates osteogenesis, particularly osteoblast differentiation, as previously described. The genomic target of RUNX2 is osteoblast-specific cis-acting element 2 (OSE2), an approximately 18-bp, highly conserved DNA sequence located in the promoter region of many osteogenic genes, such as secreted phosphoprotein 1 (SPP1), collagen 1 alpha 1 (COL1A1), alkaline phosphatase (ALP), and others [36]. Beyond the epigenetic controls of RUNX2 transcription, many miRNAs have been shown to be activators or attenuators of its expression. Lineage progression in osteoblast and chondrocyte is strictly controlled by a cell fate-determining transcription factor, RUNX2. During osteogenesis differentiation and cartilage formation, miRNAs (miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) are generally reversely expressed to RUNX2. All RUNX2 targeting miRNA (except miR-218) significantly inhibits osteoblast differentiation and can reverse its effect by corresponding anti-miRNA [37]. Attenuation of miRNA in protein translation has emerged as a crucial regulator of mesenchymal cell differentiation into osteoblast lines. The deletion of the dicer enzyme in the osteogenesis factor by Col1a1-Cre hindered the survival of the fetus after E14.5. This suggests that dicer-generated miRNA is essential for promoting prenatal osteoblast differentiation and suppressing bone development in adults [38].
Members of the miR-30 family are known as crucial modulators in osteogenic differentiation. miR-26a modulates the expression of the Smad1 protein during the osteoblastic differentiation of human adipose tissue-derived stem cells. Inhibition of miR-26a could increase osteoblast differentiation [39]. miR-135b can control osteoblast differentiation of unrestricted somatic cells (USSCs) by regulating the expression of bone-related genes. A reduction in the expression of osteogenesis markers IBSP and Osterix is known to be involved in bone mineralization in the osteogenesis of USSCs which overexpress miR-135b [40]. Exogenous miR-125b transfection inhibits osteoblast differentiation. In contrast, when endogenous miR-125b is blocked by the transfection of antisense RNA molecules, ALP activity after BMP-4 treatment is increased [41]. Both miR-141 and -200a remarkably modulated BMP-2-induced pre-osteoblast differentiation by the translational repression of Dlx5 [42]. In addition, miR-204/211 acts as a crucial negative regulator of RUNX2, which inhibits bone formation and promotes adipogenic differentiation of mesenchymal progenitor cells and BM-MSCs [43].
A few miRNAs can be promoters of bone formation. miR-218 is induced during osteoblast differentiation and has strong osteogenesis characteristics. miR-218 promotes the involvement and differentiation of bone marrow stromal cells by activating positive Wnt signaling loops [44]. miR-218 expression is promoted by the expression of the Wnt pathway gene Wnt3a, which forms an amplification circuit [45]. MiR-449a inhibits HDAC1 expression and regulates histone acetylation. Accordingly, silencing of endogenous HDAC1 expression by exogenous miR-449a maintains a histone acetylated state, stimulates RUNX2 gene expression, and rapidly promotes osteoblast differentiation [46].
miRNA-directed regulation also targets osteoclasts. Expression of miR-503, targeting RANK, is strongly reduced in circulating CD14+ macrophages of human osteoporosis patients in comparison to normal patients, and it silences miR-503 with an antagomir that promotes RANK expression and osteoclast differentiation, promotes bone resorption, and decreases bone mass in an ovariectomized mouse model [47]. MiR-223 is strongly expressed in rheumatoid arthritis (RA) synovial membrane, and excessive expression of miR-223 inhibits osteoclast formation in vitro [48]. In addition, MiR-223 expression was significantly higher in the synovial membrane of RA patients and ankle joints of collagen-induced arthritis (CIA) mice than in OA patients and normal mice. The knockdown of miR-233 by lentiviral-mediated silencing reduced the arthritis score, histological score, miR-223 expression, osteoclast formation, and bone erosion in mice with CIA [49].
Several miRNAs have recently drawn scrutiny in the bone field as potential clinical biomarkers for OVF. Ahn identified the TT genotype of miR-149aT>C and suggested it may contribute to decreased susceptibility to OVF in Korean postmenopausal women. The miR-146aCG/miR-196a2TC combined genotype and the miR-146aG/-149T/-196a2C/-449G allele combination may promote increased susceptibility to OVF. In addition, Zarecki et al. showed seven significantly (p<0.05) up-regulated miRNAs (miR-375, miR-532-3p, miR-19b-3p, miR-152-3p, miR-23a-3p, miR-335-5p, miR-21-5p) in patients with OVF [50].
2.2. Long Non-Coding (lnc) RNAs
The lncRNAs, large non-coding RNAs > 200 nucleotides in length, play crucial roles in many activities of life [51], such as epigenetic regulation, dose compensation effects, and regulation of cell differentiation (Table 3). Abnormalities of lncRNAs can cause disease, and many reports have demonstrated that lncRNAs are closely associated with the underlying mechanism in the pathogenesis of osteoporosis.
Table 3. The pathway/affected molecule or lncRNAs and their effects in bone biology.
|
lncRNA |
Pathway or Affected Molecule |
Effect on Osteoblasts and Osteoclasts |
|
lncRNA HIF1A-AS1 |
SIRT1 |
Promote the differentiation of osteoblasts |
|
LncRNA HoxA-AS3 |
EZH2, H3K27me3, RUNX2 |
Promote the differentiation of osteoblasts |
|
lncRNA MALAT1 |
miR-143, miR-204 |
Promote the differentiation of osteoblasts |
|
lncRNA MODR |
miR-454 |
Promote the differentiation of osteoblasts |
|
LncRNA KCNQ1OT1 |
miR-214 |
Promote the differentiation of osteoblasts |
|
LncRNA NTF3-5 |
miR-93-3p |
Promote the differentiation of osteoblasts |
|
LncRNA POIR |
miR-182 |
Promote the differentiation of osteoblasts |
|
LncRNA Linc-ROR |
miR-145 |
Promote the differentiation of osteoblasts |
|
LncRNA H19 |
miR-675, miR-141, miR-22 Wnt/β-catenin pathway |
Promote the differentiation of osteoblasts Inhibit the differentiation of osteoblasts |
|
LncRNA-DANCR |
EZH2, H3K27me3, RUNX2, p38 MAPK |
Inhibit the differentiation of osteoblasts |
|
LncRNA ANCR |
Wnt/β-catenin pathway |
Inhibit the differentiation of osteoblasts |
|
LncRNA HOTAIR |
Wnt/β-catenin pathway |
Inhibit the differentiation of osteoblasts |
|
lncRNA p21 |
E2, Wnt/β-catenin pathway |
Inhibit the differentiation of osteoblasts |
|
Lnc-AK045490 |
β-catenin, TCF1, LEF1, and RUNX2 |
Inhibit the differentiation of osteoblasts |
|
Lnc-AK016739 |
osteoblastic TF |
Inhibit the differentiation of osteoblasts |
|
lncRNA UCA1 |
BMP-2/(Smad1//5/8) |
Inhibit the differentiation of osteoblasts |
|
LncRNA MEG3 |
Wnt/β-catenin signaling pathway IGF1 |
Promote the differentiation of osteoclasts Inhibit the differentiation of osteoblasts |
|
LncRNA HOTAIR |
miR-17-5p, Smad7 |
Inhibit the differentiation of osteoblasts |
|
LncRNA MIAT |
miR-150-5p |
Inhibit the differentiation of osteoblasts |
|
lncRNA-ORLNC1 |
miR-296 |
Inhibit the differentiation of osteoblasts |
|
LncRNA MEG3 |
miR-133a-3p |
Inhibit the differentiation of osteoblasts |
|
LncRNA TSIX |
miR-30a-5p, and RUNX2 |
Promote the apoptosis of osteoblasts |
|
lncRNA TUG1 |
PTEN |
Promote the differentiation of osteoclasts |
|
lncRNA AK077216 |
NIP45, NFATc1 |
Promote the differentiation of osteoclasts |
|
lncRNA SNHG15 |
RANK/RANKL pathway |
Promote the differentiation of osteoclasts |
|
LncRNA-Jak3 |
NFATc1, CTSK |
Promote the differentiation of osteoclasts |
|
LncRNA LINC00311 |
DDL3 |
Promote the differentiation of osteoclasts |
|
LncRNA RP11-498C9.17 |
HDAC4 |
Inhibit the differentiation of osteoclasts |
|
LncRNA Bmncr |
RANK |
Inhibit the differentiation of osteoclasts |
|
LncRNA NONMMUT037835.2 |
RANK, NF-κB/MAPK signaling pathway |
Inhibit the differentiation of osteoclasts |
|
LncRNA-NEF |
IL-6 |
Inhibit the differentiation of osteoclasts |
HIF1A-AS (Hypoxia‑inducible factor‑1A-antisense RNA), SIRT (Sirtuin), EZH2 (enhancer of zeste homolog 2), H3K27me3 (polycomb repressive complex 2 methylates lysine 27 of histone H3-mediated trimethylation), RUNX2 (runt-related transcription factor 2), MAPK (mitogen-activated protein kinas), TCF1 (transcription factor 1), LEF1 (lymphoid enhancer-binding factor 1), TF (transcription factor), BMP (bone morphogenic protein), IGF (insulin-like growth factor), PTEN (phosphatase and tensin homolog), NIP45 ( Nuclear Factor Of Activated T Cells (NFAT) 1interacting protein), RANKL (receptor activator of nuclear factor kappa-Β ligand), CTSK (cathepsin), DDL3 (delta-like 3), HDAC4 (histone deacetylase 4), NF-κB (nuclear factor-κB), MAPK (mitogen-activated protein kinase), IL (interleukin).
Many lncRNAs have demonstrated an ability to promote osteoblast differentiation, and it has been hypothesized that they could help treat osteoporosis. The histone decarboxylase SIRT1 was shown to be a crucial positive regulator of bone mass and osteoblastogenesis. Nevertheless, the expression of SIRT1 was inversely proportional to lncRNA HIF1A-AS1 expression, which can suggest a role of lncRNA HIF1A-AS1 in osteogenic differentiation [52]. In addition, lncRNA plays a crucial role in gene regulation and is involved in a variety of cellular processes. HoxA-AS3 is increased when inducing adipogenesis of MSC and it interacts with Enhanced Of Zeste 2 (EZH2) and is required for H3K27 trimethylation of the main osteogenesis transcription factor RUNX2. As a result, lncRNA HoxA-AS3 is a crucial molecule in osteoblast differentiation [53]. In contrast to lncRNA HoxA-AS3, lncRNA-differentiation antagonizing non-protein coding RNA (DANCR) was reported to recruit EZH2 in the promotion of H3K27 trimethylation through the interaction with a 305-nt transcript and enhancer of zestehomolog2, which ultimately inhibited transcription of the target gene RUNX2 and osteogenic differentiation [54]. DANCR also modulated the proliferation and osteogenic differentiation of hBM-MSCs via the inactivation of the p38 MAPK signaling pathway [55]. The decline in anti-differentiation noncoding RNA (ANCR) accelerated the growth of periodontal ligament stem cells (PDLSC). Further, the down-regulated ANCR promotes osteogenesis differentiation of PDLSC by up-regulating the osteogenesis differentiation marker gene [56]. In addition, inhibition of lncRNAs, such as HOX transcript antisense RNA (HOTAIR), promoted ALP activity and increased the number of osteogenesis marker genes and calcified nodules in BM-MSC. However, the over-expression of HOTAIR showed the opposite effect. HOTAIR inhibited expression levels of Wntβ-catenin pathway-related proteins [57]. Furthermore, low expression of lncRNAp21 activates the Wntβ-catenin signaling pathway by increasing E2 secretion, ultimately stimulating osteogenesis, and increasing osteogenesis differentiation of MSCs in osteoporosis rat models [58]. The DKK4 gene encodes a protein that belongs to the Dickkopf family. Downregulating lncRNA H19 reduces the expression level of Dkk4, which inhibits the Wnt/β-catenin signaling pathway and negatively regulates osteogenic differentiation [59]. lncRNA AK045490 correlates with osteogenesis differentiation and strengthens skeletal tissue of mice. In the in vitro analysis of BM-MSC, AK045490 inhibited osteoblast differentiation. In vivo inhibition of AK045490 by siRNA saved bone formation in the oophorectomy osteoporosis mouse model. AK045490 inhibits nuclear dislocation of β-catenin and inhibits expression of TCF1, LEF1, and RUNX2 [60]. In a similar fashion, lncRNA AK016739 inhibits osteogenic differentiation and bone formation since it inhibits osteoblastic transcription factors [61]. By inhibiting lncRNA-UCA1, the BMP-2 (Smad158) signaling pathway in osteoblasts is activated to promote osteoblast proliferation and differentiation [62]. Moreover, the expression of serum lncRNA MEG3 in fracture patients is remarkably increased. Because LncRNA MEG3 can promote osteoblast proliferation and differentiation by activating the Wntβ-catenin signaling pathway, it is expected to become a new target for promoting fracture healing [63]. In contrast, downregulating lncRNA MEG3 inhibits osteogenic differentiation by promoting the expression of IGF1 [64]. The considerations outlined here suggest that many lncRNAs have inhibitory effects on osteogenic differentiation. As a result, silencing the expression of these specific lncRNAs using specifically targeted drugs may be possible, which may limit osteoporosis development.
lncRNAs have been reported to modulate osteoclastogenesis by modulating the expression of specific target mRNAs. Plasma lncRNA TUG1 was regulated at a higher level in patients with osteoporosis than in healthy participants. A patient with osteoporosis and a healthy patient is distinguished by increasing the regulation of plasma lncRNA TUG1. LncRNA TUG1 levels increased with advances in the clinical stage. Although excessive expression of lncRNA TUG1 accelerated proliferation and inhibited apoptosis of mouse osteoclasts, lncRNA TUG1 siRNA silencing played a reverse role [65]. The expression of lncRNA AK077216 is remarkably suppressed during osteoclast formation. Up- and down-regulation of lncRNA AK077216 promotes and inhibits osteoclast differentiation, bone absorption, and expression of related genes. lncRNA AK077216 regulates the expression of NFATc1 and promotes osteoclast formation and function [66]. Downregulation of lncRNA SNHG15 inhibits osteoclasts by modulating the RANK/RANKL pathway [67]. lncRNAs also have inhibitory effects on osteoclasts. These lncRNA expression levels were inversely correlated with osteoporosis severity. lncRNA RP11-498C9.17 is strongly correlated with many epigenetic regulatory factors, including HDAC4, HMGA1, MORF4L1, and DND1. Downregulation of HDA was reported to increase osteoclast differentiation, which suggests that lncRNA RP11-498C9.17 may modulate osteoclast production via HDAC4 signaling [68]. In addition, lncRNA Bmncr inhibits RANKL-induced osteoclast differentiation [69]. Upregulation of lncRNA NONMMUT037835.2 inhibits osteoclast differentiation, and downregulation of lncRNA-NONMMUT037835.2 promotes osteoclast formation [70]. In conclusion, these lncRNAs with effects on osteoclast differentiation may demonstrate a breakthrough in treating osteoporosis. The expression level of lncRNAs promoting osteoclast differentiation is lowered and suppressed.
3. The Possibility of Epigenetics in Treatment of Osteoporosis and OVF
The epigenetic effects on bone formation and remodeling reactions may facilitate the development of epigenetic therapeutics with the potential to treat osteoporosis and OVF. A large variety of proof-of-concept studies have demonstrated the remedial effects of miRNA and epigenetic modifiers in slowing osteoporosis development. A recombinant adeno-associated virus serotype 9 (rAAV9) can deliver artificial miRNA (amiR) to osteoclast cells of patients/animals with osteoporosis to silence expression of RANK and cathepsin K (rAAV9.amiR-rank, rAAV9.amiR-ctsk), which are major osteoporosis regulators. Because rAAV9 is very effective for the transduction of osteoclasts, systemic administration of rAAV9 with amiR rank or amiR-ctsk results in a significant increase in bone mass in mice. rAAV9.amiR-ctsk suppresses bone loss and improves bone mechanical properties in postmenopausal and senile osteoporosis mouse models [71]. A circulating miR-338 cluster in the serum could maintain bone formation capacity and increased bone mass and trabecular structure in an osteoporosis mouse model [72]. miR-672-5p induced osteoblast differentiation and mineralization in ovariectomized mice [73]. Histone methyltransferase DOT1L inhibition decreases osteoclastic activity, which can delay osteoporosis progression [74]. The knockdown of EZH2 by lentivirus-expressing shRNA rescued the abnormal fate of osteoporotic MSC. The H3K27me3 inhibitor DZNep effectively derepressed Wnt signaling and improved osteogenic differentiation of osteoporotic MSCs in vitro [75]. DNA methylation inhibitor 5-aza-20-deoxycytidine improves bone mass in disuse-induced osteopenic bone development [76].
References
- Ballane, G.; Cauley, J.A.; Luckey, M.M.; Fuleihan, G.E.-H. Worldwide prevalence and incidence of osteoporotic vertebral fractures. Osteoporos. Int. 2017, 28, 1531–1542, doi:10.1007/s00198-017-3909-3.
- Cauley, J. Epidemiology of ageing, fracture and falls. Geographic and ethnic disparities in osteoporotic fractures. Bone Abstr. 2014, 10, (6, 338–51, doi:10.1530/boneabs.3.ahp1.
- Cheng, C.; Wentworth, K.; Shoback, D.M. New Frontiers in Osteoporosis Therapy. Annu. Rev. Med. 2020, 71, 277–288, doi:10.1146/annurev-med-052218-020620.
- Hu, L.; Yin, C.; Zhao, F.; Ali, A.; Ma, J.; Qian, A. Mesenchymal Stem Cells: Cell Fate Decision to Osteoblast or Adipocyte and Application in Osteoporosis Treatment. Int. J. Mol. Sci. 2018, 19, doi:10.3390/ijms19020360.
- Richards, J.B.; Kavvoura, F.K.; Rivadeneira, F.; Styrkarsdottir, U.; Estrada, K.; Halldórsson, B.; Hsu, Y.-H.; Zillikens, M.C.; Wilson, S.G.; Mullin, B.H.; et al. Collaborative meta-analysis: Associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann. Intern. Med. 2009, 151, 528–37, doi:10.7326/0003-4819-151-8-200910200-00006.
- Ahn, T.-K.; Kim, J.-O.; Han, I.-B.; Choi, H.; Jo, M.-J.; Sohn, S.; Ropper, A.E.; Kim, N.K.; Han, I.-B. Polymorphisms of miR-146a, miR-149, miR-196a2, and miR-499 are associated with osteoporotic vertebral compression fractures in Korean postmenopausal women. J. Orthop. Res. 2017, 36, 244–253, doi:10.1002/jor.23640.
- Ahn, T.-K.; Kim, J.O.; Kim, H.W.; Park, H.S.; Shim, J.H.; Ropper, A.E.; Han, I.; Kim, N.K. 3′-UTR Polymorphisms of MTHFR and TS Associated with Osteoporotic Vertebral Compression Fracture Susceptibility in Postmenopausal Women. Int. J. Mol. Sci. 2018, 19, doi:10.3390/ijms19030824.
- Macías, I.; Alcorta-Sevillano, N.; Rodríguez, C.I.; Infante, A. Osteoporosis and the Potential of Cell-Based Therapeutic Strategies. Int. J. Mol. Sci. 2020, 21, doi:10.3390/ijms21051653.
- Sanghani-Kerai, A.; McCreary, D.; Lancashire, H.; Osagie-Clouard, L.; Coathup, M.; Blunn, G. Stem Cell Interventions for Bone Healing: Fractures and Osteoporosis. Curr. Stem Cell Res. Ther. 2018, 13, 369–377, doi:10.2174/1574888x13666180410160511.
- Schindeler, A.; McDonald, M.M.; Bokko, P.; Little, D.G. Bone remodeling during fracture repair: The cellular picture. Semin. Cell Dev. Biol. 2008, 19, 459–466, doi:10.1016/j.semcdb.2008.07.004.
- Bolander, M.E. Regulation of Fracture Repair by Growth Factors. Exp. Biol. Med. 1992, 200, 165–170, doi:10.3181/00379727-200-43410a.
- Einhorn, T.A. The Cell and Molecular Biology of Fracture Healing. Clin. Orthop. Relat. Res. 1998, 355, S7–S21, doi:10.1097/00003086-199810001-00003.
- Gerstenfeld, L.C.; Cullinane, D.M.; Barnes, G.L.; Graves, D.T.; Einhorn, T.A. Fracture healing as a post-natal developmental process: Molecular, spatial, and temporal aspects of its regulation. J. Cell. Biochem. 2003, 88, 873–884, doi:10.1002/jcb.10435.
- Ai-Aql, Z.S.; Alagl, A.S.; Graves, D.T.; Gerstenfeld, L.C.; Einhorn, T.A., Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J. Dent. Res. 2008, 87, 107–118.
- Giannoudis, P.V.; Einhorn, T.A.; Marsh, D. Fracture healing: The diamond concept. Injury 2007, 38, S3–S6, doi:10.1016/s0020-1383(08)70003-2.
- Stone, C. A molecular approach to bone regeneration. Br. J. Plast. Surg. 1997, 50, 369–373, doi:10.1016/s0007-1226(97)90547-6.
- Barnes, G.L.; Kostenuik, P.J.; Gerstenfeld, L.C.; A Einhorn, T. Growth Factor Regulation of Fracture Repair. J. Bone Miner. Res. 1999, 14, 1805–1815, doi:10.1359/jbmr.1999.14.11.1805.
- Khosla, S.; Riggs, B.L. Pathophysiology of Age-Related Bone Loss and Osteoporosis. Endocrinol. Metab. Clin. North. Am. 2005, 34, 1015–1030, doi:10.1016/j.ecl.2005.07.009.
- Baccarelli, A.A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251, doi:10.1097/mop.0b013e32832925cc.
- Yasui, T.; Hirose, J.; Aburatani, H.; Tanaka, S. Epigenetic regulation of osteoclast differentiation. Ann. New York Acad. Sci. 2011, 1240, 7–13, doi:10.1111/j.1749-6632.2011.06245.x.
- Kurotaki, D.; Yoshida, H.; Tamura, T. Epigenetic and transcriptional regulation of osteoclast differentiation. Bone 2020, 138, doi:10.1016/j.bone.2020.115471.
- Guil, S.; Esteller, M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009, 41, 87–95, doi:10.1016/j.biocel.2008.09.005.
- Feinberg, A.P. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. New Engl. J. Med. 2018, 378, 1323–1334, doi:10.1056/nejmra1402513.
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92, doi:10.1038/nrg.2017.80.
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M., The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489.
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589, doi:10.1038/s41580-019-0143-1.
- Schübeler, D. Function and information content of DNA methylation. Nat. Cell Biol. 2015, 517, 321–326, doi:10.1038/nature14192.
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451, doi:10.1007/s00018-018-2940-7.
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J., Targeting noncoding RNAs in disease. J. Clin. Invest. 2017, 127, 761–771.
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418, doi:10.1038/s41591-019-0376-8.
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, aad9780, doi:10.1126/science.aad9780.
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134, doi:10.1038/nm.4036.
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101, doi:10.1038/nrm.2016.140.
- Pratt, A.J.; Macrae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901, doi:10.1074/jbc.r900012200.
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933, doi:10.4161/rna.24604.
- Cohen, M.M., Jr. Perspectives on RUNX genes: An update. Am. J. Med. Genet. A 2009, 149, 2629–2646.
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor RUNX2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868.
- Gaur, T.; Hussain, S.; Mudhasani, R.; Parulkar, I.; Colby, J.L.; Frederick, D.; Kream, B.E.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 2010, 340, 10–21, doi:10.1016/j.ydbio.2010.01.008.
- Luzi, E.; Marini, F.; Sala, S.C.; Tognarini, I.; Galli, G.; Brandi, M.L. Osteogenic Differentiation of Human Adipose Tissue-Derived Stem Cells Is Modulated by the miR-26a Targeting of the SMAD1 Transcription Factor. J. Bone Miner. Res. 2007, 23, 287–295, doi:10.1359/jbmr.071011.
- Schaap-Oziemlak, A.M.; Raymakers, R.A.; Bergevoet, S.M.; Gilissen, C.; Jansen, B.J.; Adema, G.J.; Kogler, G.; le Sage, C.; Agami, R.; van der Reijden, B.A.; et al. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010, 19, 877–885.
- Mizuno, Y.; Yagi, K.; Tokuzawa, Y.; Kanesaki-Yatsuka, Y.; Suda, T.; Katagiri, T.; Fukuda, T.; Maruyama, M.; Okuda, A.; Amemiya, T.; et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008, 368, 267–272, doi:10.1016/j.bbrc.2008.01.073.
- Itoh, T.; Nozawa, Y.; Akao, Y. MicroRNA-141 and -200a Are Involved in Bone Morphogenetic Protein-2-induced Mouse Pre-osteoblast Differentiation by Targeting Distal-less Homeobox 5. J. Biol. Chem. 2009, 284, 19272–19279, doi:10.1074/jbc.m109.014001.
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 Regulates Runx2 Protein Expression and Mesenchymal Progenitor Cell Differentiation. STEM CELLS 2009, 28, 357–364, doi:10.1002/stem.288.
- Hassan, M.Q.; Maeda, Y.; Taipaleenmaki, H.; Zhang, W.; Jafferji, M.; Gordon, J.A.R.; Li, Z.; Croce, C.M.; Van Wijnen, A.J.; Stein, J.L.; et al. miR-218 Directs a Wnt Signaling Circuit to Promote Differentiation of Osteoblasts and Osteomimicry of Metastatic Cancer Cells. J. Biol. Chem. 2012, 287, 42084–42092, doi:10.1074/jbc.m112.377515.
- Zhang, W.B.; Zhong, W.J.; Wang, L. A signal-amplification circuit between miR-218 and Wnt/beta-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone 2014, 58, 59–66.
- Liu, T.; Hou, L.; Zhao, Y.; Huang, Y. Epigenetic silencing of HDAC1 by miR-449a upregulates Runx2 and promotes osteoblast differentiation. Int. J. Mol. Med. 2014, 35, 238–246, doi:10.3892/ijmm.2014.2004.
- Chen, C.; Cheng, P.; Xie, H.; Zhou, H.-D.; Wu, X.-P.; Liao, E.-Y.; Luo, X. MiR-503 Regulates Osteoclastogenesis via Targeting RANK. J. Bone Miner. Res. 2014, 29, 338–347, doi:10.1002/jbmr.2032.
- Shibuya, H.; Nakasa, T.; Adachi, N.; Nagata, Y.; Ishikawa, M.; Deie, M.; Suzuki, O.; Ochi, M. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod. Rheumatol. 2013, 23, 674–685.
- Li, Y.-T.; Chen, S.-Y.; Wang, C.-R.; Liu, M.-F.; Lin, C.-C.; Jou, I.-M.; Shiau, A.-L.; Wu, C.-L. Brief Report: Amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2012, 64, 3240–3245, doi:10.1002/art.34550.
- Zarecki, P.; Hackl, M.; Grillari, J.; Debono, M.; Eastell, R. Serum microRNAs as novel biomarkers for osteoporotic vertebral fractures. Bone 2019, 130, doi:10.1016/j.bone.2019.115105.
- Zhao, K.; Zhao, Q.; Guo, Z.; Chen, Z.; Hu, Y.; Su, J.; Chen, L.; He, Z.; Cai, X.; Chen, M.; et al. Hsa_Circ_0001275: A Potential Novel Diagnostic Biomarker for Postmenopausal Osteoporosis. Cell. Physiol. Biochem. 2018, 46, 2508–2516, doi:10.1159/000489657.
- Huynh, N.P.T.; Anderson, B.A.; Guilak, F.; McAlinden, A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect. Tissue Res. 2017, 58, 116–141, doi:10.1080/03008207.2016.1194406.
- Zhu, X.X.; Yan, Y.W.; Chen, D.; Ai, C.Z.; Lu, X.; Xu, S.S.; Jiang, S.; Zhong, G.S.; Chen, D.B.; Jiang, Y.Z. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016, 7, 63561–63570.
- Zhang, L.; Zhang, P.; Sun, X.; Zhou, L.; Zhao, J. Long non-coding RNA DANCR regulates proliferation and apoptosis of chondrocytes in osteoarthritis via miR-216a-5p-JAK2-STAT3 axis. Biosci. Rep. 2018, 38, doi:10.1042/bsr20181228.
- Peng, S.; Cao, L.; He, S.; Zhong, Y.; Ma, H.; Zhang, Y.; Shuai, C. An Overview of Long Noncoding RNAs Involved in Bone Regeneration from Mesenchymal Stem Cells. Stem Cells Int. 2018, 2018, 1–11, doi:10.1155/2018/8273648.
- Jia, Q.; Jiang, W.; Ni, L.,Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch. Oral Biol. 2015, 60, 234–241.
- Shen, J.J.; Zhang, C.H.; Chen, Z.W.; Wang, Z.X.; Yang, D.C.; Zhang, F.L.; Feng, K.H. LncRNA HOTAIR inhibited osteogenic differentiation of BMSCs by regulating Wnt/beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7232–7246.
- Yang, K.; Tian, N.; Liu, H.; Tao, X.Z.; Wang, M.X.; Huang, W., LncRNAp21 promotes osteogenic differentiation of mesenchymal stem cells in the rat model of osteoporosis by the Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4303–4309.
- Li, B.; Liu, J.; Zhao, J.; Ma, J.X.; Jia, H.B.; Zhang, Y.; Xing, G.S.; Ma, X.L. LncRNA-H19 Modulates Wnt/beta-catenin Signaling by Targeting Dkk4 in Hindlimb Unloaded Rat. Orthop. Surg. 2017, 9, 319–327.
- Li, D.; Tian, Y.; Yin, C.; Huai, Y.; Zhao, Y.; Su, P.; Wang, X.; Pei, J.; Zhang, K.; Yang, C.; et al. Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via beta-Catenin/TCF1/RUNX2 Signaling Axis. Int. J. Mol. Sci. 2019, 20.
- Yin, C.; Tian, Y.; Yu, Y.; Wang, H.; Wu, Z.; Huang, Z.; Zhang, Y.; Li, D.; Yang, C.; Wang, X.; et al. A novel long noncoding RNA AK016739 inhibits osteoblast differentiation and bone formation. J. Cell. Physiol. 2019, 234, 11524–11536, doi:10.1002/jcp.27815.
- Zhang, R.-F.; Liu, J.-W.; Yu, S.-P.; Sun, D.; Wang, X.-H.; Fu, J.-S.; Xie, Z. LncRNA UCA1 affects osteoblast proliferation and differentiation by regulating BMP-2 expression. Eur Rev. Med. Pharmacol Sci 2019, 23, 6774–6782.
- Li, X.G.; Liu, S.C.; Qiao, X.F.; Kong, Y.; Liu, J.G.; Peng, X.M.; Wang, Y.X.; Abdulkarim Mohammed Al-Mohana, R.A. LncRNA MEG3 promotes proliferation and differentiation of osteoblasts through Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4521–4529.
- Liu, Y.; Liu, C.; Zhang, A.; Yin, S.; Wang, T.; Wang, Y.; Wang, M.; Liu, Y.; Ying, Q.; Sun, J.; et al. Down-regulation of long non-coding RNA MEG3 suppresses osteogenic differentiation of periodontal ligament stem cells (PDLSCs) through miR-27a-3p/IGF1 axis in periodontitis. Aging 2019, 11, 5334–5350, doi:10.18632/aging.102105.
- Han, Y.; Liu, C.; Lei, M.; Sun, S.; Zheng, W.; Niu, Y.; Xia, X. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J. Orthop. Surg. Res. 2019, 14, 416.
- Liu, C.; Cao, Z.; Bai, Y.; Dou, C.; Gong, X.; Liang, M.; Dong, R.; Quan, H.; Li, J.; Dai, J.; et al. LncRNA AK077216 promotes RANKL-induced osteoclastogenesis and bone resorption via NFATc1 by inhibition of NIP45. J. Cell. Physiol. 2019, 234, 1606–1617, doi:10.1002/jcp.27031.
- Liu, Z.-Z.; Zhang, C.-Y.; Huang, L.-L.; Liu, W. Elevated expression of lncRNA SNHG15 in spinal tuberculosis: Preliminary results. Eur Rev. Med. Pharmacol Sci 2019, 23, 9017–9024.
- Li, L.; Wang, X.Q.; Liu, X.T.; Guo, R.; Zhang, R.D. Integrative analysis reveals key mRNAs and lncRNAs in monocytes of osteoporotic patients. Math. Biosci. Eng. 2019, 16, 5947–5970, doi:10.3934/mbe.2019298.
- Chen, R.-S.; Zhang, X.-B.; Zhu, X.-T.; Wang, C.-S. LncRNA Bmncr alleviates the progression of osteoporosis by inhibiting RANML-induced osteoclast differentiation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9199–9206.
- Chang, Y.; Yu, D.; Chu, W.; Liu, Z.; Li, H.; Zhai, Z. LncRNA expression profiles and the negative regulation of lncRNA-NOMMUT037835.2 in osteoclastogenesis. Bone 2020, 130, doi:10.1016/j.bone.2019.115072.
- Yang, Y.-S.; Xie, J.; Chaugule, S.; Wang, D.; Kim, J.-M.; Kim, J.; Tai, P.W.; Seo, S.-K.; Gravallese, E.; Gao, G.; et al. Bone-Targeting AAV-Mediated Gene Silencing in Osteoclasts for Osteoporosis Therapy. Mol. Ther. Methods Clin. Dev. 2020, 17, 922–935, doi:10.1016/j.omtm.2020.04.010.
- Lin, C.; Yu, S.; Jin, R.; Xiao, Y.; Pan, M.; Pei, F.; Zhu, X.; Huang, H.; Zhang, Z.; Chen, S.; et al. Circulating miR-338 Cluster activities on osteoblast differentiation: Potential Diagnostic and Therapeutic Targets for Postmenopausal Osteoporosis. Theranostics 2019, 9, 3780–3797, doi:10.7150/thno.34493.
- Ahmad, N.; Kushwaha, P.; Karvande, A.; Tripathi, A.K.; Kothari, P.; Adhikary, S.; Khedgikar, V.; Mishra, V.K.; Trivedi, R. MicroRNA-672-5p Identified during Weaning Reverses Osteopenia and Sarcopenia in Ovariectomized Mice. Mol. Ther. Nucleic Acids 2019, 14, 536–549, doi:10.1016/j.omtn.2019.01.002.
- Gao, Y.; Ge, W. The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis. Cell Death Dis. 2018, 9, 1–16, doi:10.1038/s41419-017-0040-5.
- Jing, H.; Liao, L.; An, Y.; Su, X.; Liu, S.; Shuai, Y.; Zhang, X.; Jin, Y. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Mol. Ther. 2016, 24, 217–229, doi:10.1038/mt.2015.152.
- Li, B.; Zhao, J.; Ma, J.-X.; Li, G.-M.; Zhang, Y.; Xing, G.-S.; Liu, J.; Ma, X.-L. Overexpression of DNMT1 leads to hypermethylation of H19 promoter and inhibition of Erk signaling pathway in disuse osteoporosis. Bone 2018, 111, 82–91, doi:10.1016/j.bone.2018.03.017.