Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ayman Elbehiry | -- | 2658 | 2022-11-25 07:58:46 | | | |
| 2 | Dean Liu | -4 word(s) | 2654 | 2022-11-28 03:40:34 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Elbehiry, A.; Aldubaib, M.; Abalkhail, A.; Marzouk, E.; Albeloushi, A.; Moussa, I.; Ibrahem, M.; Albazie, H.; Alqarni, A.; Anagreyyah, S.; et al. Implications of MALDI-TOF MS for Microbial Recognition. Encyclopedia. Available online: https://encyclopedia.pub/entry/36514 (accessed on 08 March 2026).
Elbehiry A, Aldubaib M, Abalkhail A, Marzouk E, Albeloushi A, Moussa I, et al. Implications of MALDI-TOF MS for Microbial Recognition. Encyclopedia. Available at: https://encyclopedia.pub/entry/36514. Accessed March 08, 2026.
Elbehiry, Ayman, Musaad Aldubaib, Adil Abalkhail, Eman Marzouk, Ahmad Albeloushi, Ihab Moussa, Mai Ibrahem, Hamad Albazie, Abdullah Alqarni, Sulaiman Anagreyyah, et al. "Implications of MALDI-TOF MS for Microbial Recognition" Encyclopedia, https://encyclopedia.pub/entry/36514 (accessed March 08, 2026).
Elbehiry, A., Aldubaib, M., Abalkhail, A., Marzouk, E., Albeloushi, A., Moussa, I., Ibrahem, M., Albazie, H., Alqarni, A., Anagreyyah, S., Alghamdi, S., & Rawway, M. (2022, November 25). Implications of MALDI-TOF MS for Microbial Recognition. In Encyclopedia. https://encyclopedia.pub/entry/36514
Elbehiry, Ayman, et al. "Implications of MALDI-TOF MS for Microbial Recognition." Encyclopedia. Web. 25 November, 2022.
Copy Citation
MALDI-TOF MS has various benefits over the conventional method of biochemical identification, including ease of use, speed, accuracy, and low cost.
MALDI-TOF MS
healthcare settings
microbial identification
1. Bacterial Identification
The most common and best-optimized utilization of MALDI-TOF to date is for the regular recognition of clinical pathogens. Because of its incredible rapidity for microbial detection, whether from cultivation or directly from medical specimens, MALDI-TOF has during the past two decades become the standard method. The majority of microbial pathogens frequently seen in clinical microbiology facilities may now be successfully identified, employing MALDI-TOF due to an increase in the technique’s reliability over time [1][2]. The ability of MALDI-TOF to correctly identify species that are members of the Enterobacteriaceae (e.g., Salmonella spp. and Cronobacter spp.) at the genus level, according to a collaborative study completed in 2018 [3]. Aeromonas, Citrobacter, Escherichia, Plesiomonas, Providencia, Serratia, and Yersinia, also closed-related genera, were investigated in the same study, allowing the scientists to exclude Salmonella or Cronobacter confusion. It has been widely shown that MALDI-TOF can properly detect the most prevalent species of the Enterobacteriaceae [4][5]. Even so, it has presumably been revealed that MALDI-TOF misidentifies Shigella species as Escherichia coli and only partially distinguishes between those that belong to the Citrobacter freundii, Enterobacter cloacae, and Salmonella enterica species [4][6].
The practice of the majority of the microbiology labs currently includes the detection of non-fermenting Gram-negative rods by MALDI Biotyper. Various MALDI-TOF methods have been proven to properly identify the most frequent species seen in clinical settings (e.g., Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Moraxella catharralis) in 93.6–96.6% of the cases studied [1][7]. However, as a result of the similarity among the species in this genus, certain species have only been recognized on a broad scale (e.g., Burkholderia cepacia and Acinetobacter baumannii) or at a genus level (e.g., Achromobacter species, Chryseobacter species, and Ralstonia species). These species pose a problem for DNA-based techniques as well, but recognition at the species level is typically not important from a medical viewpoint. Updates in the library are recommended, utilizing reference strains accurately described by genetic approaches where recognition at the species level is necessary to determine the antimicrobial activities. Nevertheless, subsequent datasets and spectral analyzing advancements may make achieving a higher degree of reliability simpler [1].
When supplemented datasets are used, MALDI-TOF has also proved successful in identifying various Gram-negative bacteria, such as the HACEK group (Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, Kingella), with rates of 100% species precision [8]. In one investigation, a commercialized dataset produced 66.0% appropriate species level recognition but 93.0% proper genus level. This also applies to the Neisseria genus, because several species that are included in the normal microbial community have been mistaken for Neisseria meningitides [9]. The only way to avoid these mistakes and guarantee a correct assessment is through the usage of current and extended databases. As demonstrated in another category, the level of reliability presently attained for the recognition of Gram-negative organisms via MALDI-TOF is still astonishing given that it is carried out in less than 1 h [10]. Oviaño and his colleagues [10] conducted a comparison study assessing the detection of suspected colonies from fecal samples by MBT compass software vs. conventional morphological approaches. They discovered that the full MALDI identification process, from preparation of the sample to obtaining the final result, was finished in less than 30 min, cutting the test’s delivery time by two to three days as described in another study [11].
Gram-positive pathogens have consistently been difficult for MALDI-TOF to analyze. Some of them are challenging to distinguish because of their complicated composition of cell walls and the tight kinship between the species within the major genera. The success rate of identifying these pathogens at the species level has significantly risen as a result of the application of formic acid for a quick, on-plate protein extraction [12]. Despite significant advances in the databases that are provided, it is occasionally simple to distinguish between various Streptococcus species. An effort has been made to identify particular peaks that enable reliable distinction [13]. In addition, a method that uses the top 10 identifications obtained by MALDI-TOF has been created with the same objective [14]. With regard to anaerobes, MALDI-TOF has demonstrated to offer species-level identification compared with genetic methods in a quick and economical manner [15]. According to a multicentric study called ENRIA, which enabled the enhancement of the MBT database with approximately 6903 bacterial species and more than 60 distinct genera, significant advancements have been made in the accurate identification of anaerobes [16]. The upgraded database had a particularly big influence on the bacterial detection of Gram-positive anaerobic isolates, of which recognition was considered to be difficult due to the absence of spectral data from this category of microorganisms in earlier iterations of commercialized databases [1].
Mycobacteria are commonly recognized by MALDI TOF MS technology [17][18]. Unfortunately, this technology has the same drawback as real-time PCR in that it cannot distinguish between various Mycobacterium tuberculosis complex (MBTC) species [19]. Therefore, it is now unable to specifically identify tuberculosis species with this technology [20]. Since the advent of up-to-date, thorough datasets and the development of streamlined sample processing techniques in recent years, MALDI-TOF MS has been able to differentiate between a growing number of non-tuberculous Mycobacterium species. While MALDI-TOF MS is still able to differentiate between MBTC species in the coming year, this is still one of its greatest challenges [18]. It was demonstrated that the in vitro diagnostic method created by the company of Bruker Daltonics (Bremen, Germany), that uses crystal particles to mechanically interrupt the microbial biomass accompanied by ribosomal extraction, can provide identification at the species level [1]. Species with complex morphologies, for instance the Mycobacterium avium, can be successfully differentiated via a species-specific marker [21]. Overall, MALDI-TOF enables extremely accurate detection of Mycobacteria that are not tuberculous. The precision of this authentication is comparable to that offered by molecular biology techniques, although MALDI-TOF requires a response time of about one hour since the specimens must be heat-inactivated for half an hour [22]. Similar to mycobacteria, Nocardia and Streptomyces belonging to the order Actinomycetales must have their cell walls damaged in order for MALDI-TOF to correctly identify them. Because of their complex nomenclature, it can be difficult to differentiate between similar species.
2. Yeasts and Filamentous Fungi Identification
There are two major MALDI-TOF MS systems on the market today: the MBT (Bruker Daltonics) and the Vitek MS (BioMerieux). Although a distinct fungi database only became accessible for mold detection for experimental use in 2012, the FDA only approved the MBT in 2013 for the identification of bacteria and yeast. For bacteria and yeast identification, the FDA only approved the MBT in 2013 after a separate fungi database had been made available for experimental use in 2012. The FDA, however, approved the Vitek MS version 3.0 in 2017 with a library that included more bacteria, yeasts, mold, and mycobacteria [23]. In 2017, the FDA approved a version of Vitek MS containing additional bacteria, yeasts, molds, and mycobacteria. In spite of the fact that chances of success might vary greatly depending on microorganism type, database version, extraction process conditions, and threshold level, scientists have observed that both MBT and VITEK MS systems are capable of fairly excellent recognition of yeast [23]. In clinical microbiology labs nowadays, MALDI-TOF is applied routinely for the discovery of yeast species [24]. The identification of yeasts, particularly those from regularly occurring genera (e.g., Cryptococcus and Candida), is performed immediately using cultivated colonies on growth media and from medical samples, despite the fact that the utilize of protein extraction was recorded in earlier investigations. To identify non-Candida genera, most researchers recommend using internal databases.
Similarly, fungal species have been verified in the same way that nontuberculous mycobacteria have been verified: verification of these microbes usually requires prior disturbance to penetrate their cellular structure, the complexity of fungi is higher than that of bacteria, and the structure of protein molecules has changed over time as well. Until relatively recently, there were no comprehensive commercial databases to address these challenges. It is therefore not surprising to see a number of authors building their own internal libraries as a result of certain sample preparations [25]. There is currently no accepted standard procedure for detecting fungal species, but researchers working on MALDI-TOF have used equivalent mixtures of mechanical and chemical methods to solve this problem [26]. Eventually, Normand et al. [27] conducted a more extensive effort. To compare spectra from unidentified fungal isolates, a library with approximately 11,000 stored spectra from about 938 filamentous fungi of clinical significance has been created and made available on the web. Aspergillus, Trichophyton, and Microsporum species that are clinically meaningful can be correctly identified using the library, which enables microbiology labs to quickly and accurately carry out the necessary fungal identification [28][29].
3. Viral Identification
Numerous studies have investigated the use of MALDI-TOF MS to detect a wide variety of viruses, including Papillomaviridae, Picornavirus, and Orthomyxoviridae families [30]. As reported by Sjöholm et al. [31], MALDI-TOF MS had a reliability rate of 95.6% and a specificity of 98.0% when analyzing 882 fluid samples. With the aid of MALDI-TOF MS, Cai and co-workers developed a high-throughput genetic analysis method with a matching rate of 80.1% (285/356). Based on genetic analysis and MALDI-TOF MS, Peng et al. identified enteroviruses and 93.4% (225/241) of the findings were in agreement [32]. A MALDI-TOF MS analysis was also conducted to distinguish different influenza virus types, and the findings were 100% (29/29) compatible with genetic analyses [33]. To detect other respiratory viruses, such as influenza, in cell cultures of clinical samples, MALDI-TOF MS is used to analyze the protein spectral composition of the coat proteins of intact tobacco mosaic viruses [34][35][36]. The SARS-CoV-1 proteins and trypsin-digested peptides were additionally identified by MALDI-TOF MS [37]. This tool developed by Yoshinari and his coworkers combines simple purification processes with a rapid recognition phase to identify SARS-CoV-2-specific peptides accurately in nasopharyngeal swabs, which may offer a more accurate method of diagnosing COVID-19 in the future [34]. For COVID-19 testing, MALDI-TOF MS has been suggested as a viable approach [36][38]. Lazari and coworkers used MALDI-TOF protein signatures from salivary swabs to develop a new strategy for diagnosing COVID-19 and demonstrated that this technique could identify and predict the virus [39].
In the near future, clinical virology will benefit from the detection of resistant protein markers, the determinants of virulence, and MALDI-TOF MS applications for more direct detection of clinical samples. A change in working procedures would, however, be necessary if this technology were routinely used for virology diagnosis [40]. The fact that sensitivity can be identified, among other technical advancements, requires an increased level of integration into laboratory workflows. Diagnostics can be made immediately from clinical samples, and microorganism databases need to be updated continuously. MALDI-TOF should become available for everyday viral diagnostic testing as technological advancement plays a key role in viral identification through MS techniques.
4. Detection of Antibiotic Resistance
An urgent need exists for creative solutions to the rising rates of multidrug-resistant bacteria, not simply for treatment and preventive measures but also for the identification and characterization of isolates that are resistant to antibiotics [41][42]. The main barriers to starting a prompt targeted antibiotic therapy and/or infection prevention and control measures are the difficulty of conventional morphological sensitivity testing protocols and the limitations of quick DNA-based molecular diagnostics. An improved therapeutic results depend on receiving early, adequate treatment [43]. The benchmark standards for phenotypic antibiotic susceptibility testing (AST) depends on the identification of bacterial activity or suppression in the presence of an antibiotic. The currently offered conventional techniques of assessing AST take roughly a day to generate a result, despite the fact that AST seems to be most important since it can detect all types of resistance regardless of the mechanisms underlying that resistance [44].
New fast phenotypic AST methods are therefore required. Here, researchers introduce a freshly created technique known as a straightforward microdroplet growth test for the quick discovery of antibiotic resistance based on MALDI technology. In spite of the microbiological species, antimicrobial drug in issue, or fundamental resistance strategy, this method enables comprehensive morphologic AST, which enables quick separation between resistant and sensitive isolates [42]. Due to MALDI-TOF MS’s adaptability, increasing numbers of applications relying on this approach are beginning to appear. The identification of virulent biomarkers and the screening of antibiotic resistance are among the most intriguing ones currently being used or soon to be used in clinical settings [1]. Recently, MALDI-TOF MS is considered one of the most common methods used detecting antimicrobial resistance through measuring the hydrolysis of different antibiotics such as beta-lactamases and carbapenemases [45]. The beta-lactamase activities are conducted in a relatively similar manner across all the research papers [1]. An antibiotic buffer is reconstituted in fresh bacterial culture and incubated at 37 °C with stirring for one to two hours. As soon as the culture period is over, the mixture is spun, and the supernatant is transferred to a MALDI-TOF work-piece to be analyzed. Samples can be inserted, dehydrated, and then analyzed by MALDI-TOF MS. Researchers have reported that cephalosporins have the spectra of cefotaxime, ceftazidime, ceftriaxone, cefpodoxime, and cefepime as well as their hydrolysis products [10]. Because of cefepime’s limited sensitivity, which causes false positive results in the hydrolysis, it has been rejected for MALDI-TOF identification of beta-lactamases, while ceftriaxone is the most reactive, selective, and fastest [1].
Extended spectrum beta-lactactamases (ESBL) producing bacteria would exhibit positive degradation after 30 min of incubation (Figure 1). Although cefotaxime has more expertise, the outcomes for cefotaxime and cefpodoxime are comparable. Both have susceptibility levels exceeding 90%, however for both, a 1 h incubation period is advised. Ceftazidime is not advised as a drug of choice for the investigation of beta-lactactamases since it significantly reduces the efficiency of hydrolysis and, subsequently, MALDI-TOF MS recognition. The suggested incubation period of three hours is based on the susceptibility being less than 90% [1].
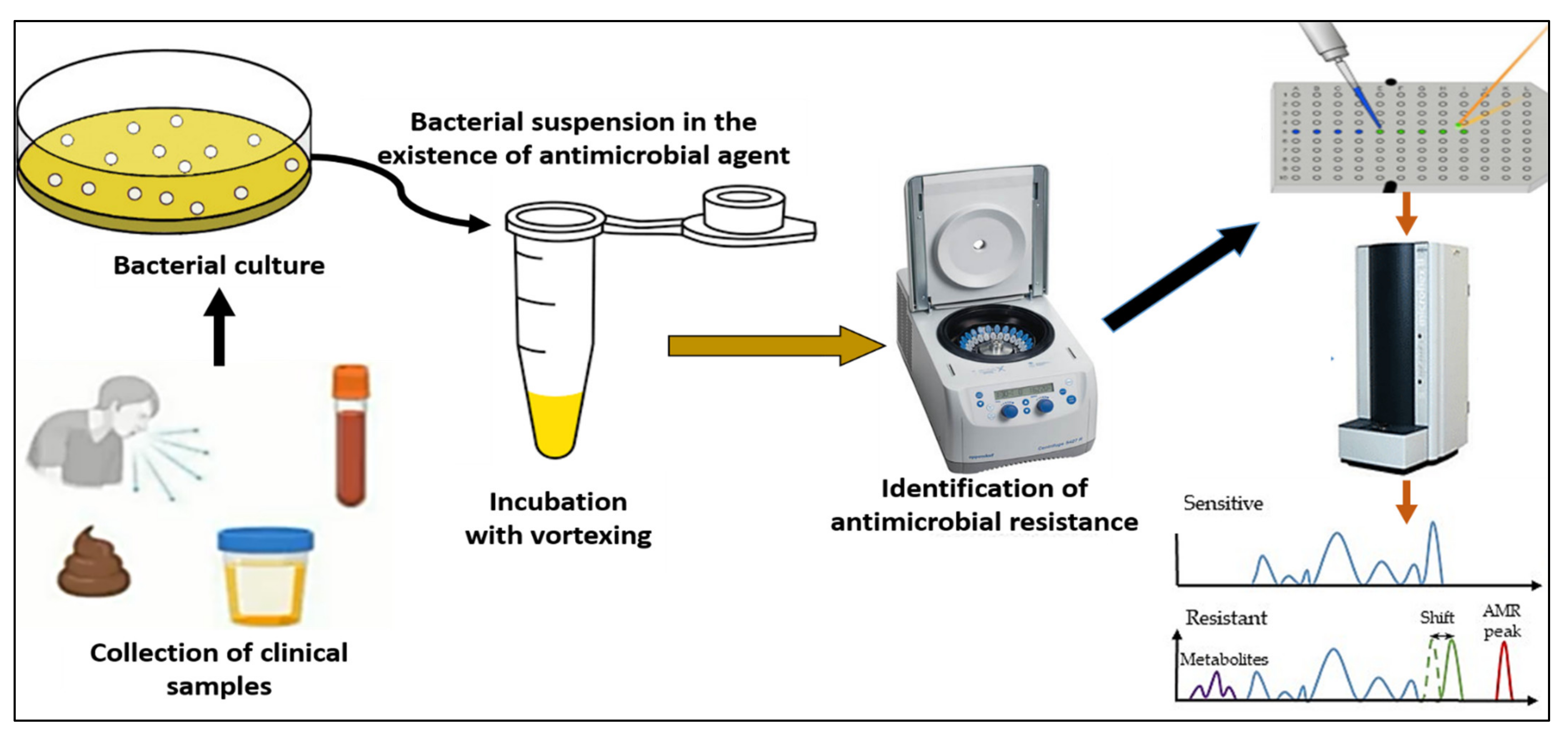
Figure 1. A workflow for MALDI-TOF detection of antimicrobial resistance.
With the aid of MS, it has been described how ertapenem, meropenem and imipenem are hydrolyzed in the context of carbapenem antibiotics. A 2 h incubation period is recommended for both ertapenem and meropenem due to their high sensitivity. Mass spectra of ertapenem show clearly defined peaks, however mass spectra of meropenem and imipenem are excluded [46]. It has been suggested that dihydroxybenzoic acid could be used as a matrix for analyzing meropenem and its hydrolyzed product [47]. The MBT STAR-BL IVD software, used in conjunction with the MBT STAR-Carba IVD Kit and the MBT STAR-Cepha IVD Kit, is a tolerance package for the MBT. The program automatically analyzes peaks and produces a merged report including the verification of the strain of bacteria and its imipenem sensitivity. Using this technology, even non-MS specialists can evaluate hydrolysis as it is automated [1]. In order to differentiate sensitive from resistant bacteria of the same species, the peak evaluation method is used. The peaks can be allied with a particular design if the transcription of a protein or peptide is related to the resistance phenotype directly or indirectly. By comparing mass peaks, the user can determine which germs are vulnerable and which ones are resistant. Using the spectra of beta-lactamase as a marker, Camara and Hays [48] distinguished between ampicillin-resistant strains of plasmid-transformed E. coli from different sources. MALDI-TOF MS was successfully used in 2011 to distinguish between Bacteroides fragilis that are cfiA-positive and cfiA-negative, and subsequently their carbapenem resistance. In spite of this, these examinations still seem to suggest additional incubation time, albeit less than for antibiograms, which intrinsically delays assessment of which antibiotherapy will work best. Before MALDI-TOF MS can be regarded as the new reference approach for the screening of antibiotic resistance in regular investigations, there remains a great deal of work concerning the review on the identification of antimicrobial resistance by specialized MALDI-TOF spectra patterns. There are still a lot of unanswered problems, thus further research on pathogenic organisms is needed. The scientists need to be very interested in research on critically important antibiotics like cephalosporins or macrolides, which are frequently utilized to treat infections in hospital settings despite the lack of evidence on them. Last but not least, the combined strategy of molecular and MALDI-TOF MS should quickly replace other methods as the primary method for the most accurate and quick diagnosis of antimicrobial resistance in clinical microbiology labs.
References
- Oviaño, M.; Rodríguez-Sánchez, B. MALDI-TOF mass spectrometry in the 21st century clinical microbiology laboratory. Enferm. Infecc. Y Microbiol. Clin. 2021, 39, 192–200.
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’Hom, G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554.
- Bastin, B.; Bird, P.; Benzinger, M.J.; Crowley, E.; Agin, J.; Goins, D.; Sohier, D.; Timke, M.; Shi, G.; Kostrzewa, M. Confirmation and identification of Salmonella spp., Cronobacter spp., and other Gram-negative organisms by the Bruker MALDI biotyper method: Collaborative study, first action 2017.09. J. AOAC Int. 2018, 101, 1593–1609.
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551.
- Martiny, D.; Busson, L.; Wybo, I.; El Haj, R.A.; Dediste, A.; Vandenberg, O. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 1313–1325.
- Rodrigues, C.; Passet, V.; Rakotondrasoa, A.; Brisse, S. Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and related phylogroups by MALDI-TOF mass spectrometry. Front. Microbiol. 2018, 9, 3000.
- Alby, K.; Gilligan, P.H.; Miller, M.B. Comparison of matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry platforms for the identification of Gram-negative rods from patients with cystic fibrosis. J. Clin. Microbiol. 2013, 51, 3852–3854.
- Couturier, M.R.; Mehinovic, E.; Croft, A.C.; Fisher, M.A. Identification of HACEK clinical isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 1104–1106.
- Hong, E.; Bakhalek, Y.; Taha, M.-K. Identification of Neisseria meningitidis by MALDI-TOF MS may not be reliable. Clin. Microbiol. Infect. 2019, 25, 717–722.
- Oviaño, M.; Gómara, M.; Barba, M.J.; Revillo, M.J.; Barbeyto, L.P.; Bou, G. Towards the early detection of β-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J. Antimicrob. Chemother. 2017, 72, 2259–2262.
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791.
- Rodrıguez-Sánchez, B.; Marın, M.; Sánchez-Carrillo, C.; Cercenado, E.; Ruiz, A.; Rodrıguez-Créixems, M.; Bouza, E. Improvement of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of difficult-toidentify bacteria and its impact in the workflow of a clinical microbiology laboratory. Diagn. Microbiol. Infect. Dis. 2014, 79, 1–6.
- Marín, M.; Cercenado, E.; Sánchez-Carrillo, C.; Ruiz, A.; González, Á.G.; Rodríguez-Sánchez, B.; Bouza, E. Accurate differentiation of Streptococcus pneumoniae from other species within the Streptococcus mitis group by peak analysis using MALDI-TOF MS. Front. Microbiol. 2017, 8, 698.
- Harju, I.; Lange, C.; Kostrzewa, M.; Maier, T.; Rantakokko-Jalava, K.; Haanperä, M. Improved differentiation of Streptococcus pneumoniae and other S. mitis group streptococci by MALDI Biotyper using an improved MALDI Biotyper database content and a novel result interpretation algorithm. J. Clin. Microbiol. 2017, 55, 914–922.
- Rodríguez-Sánchez, B.; Alcalá, L.; Marín, M.; Ruiz, A.; Alonso, E.; Bouza, E. Evaluation of MALDI-TOF MS (matrix-assisted laser desorption-ionization time-of-flight mass spectrometry) for routine identification of anaerobic bacteria. Anaerobe 2016, 42, 101–107.
- Veloo, A.; Jean-Pierre, H.; Justesen, U.; Morris, T.; Urban, E.; Wybo, I.; Kostrzewa, M.; Friedrich, A.; Shah, H.; Nagy, E. Validation of a for anaerobic bacteria optimized MALDI-TOF MS biotyper database: The ENRIA project. Anaerobe 2018, 54, 224–230.
- Patel, R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111.
- Alcaide, F.; Amlerová, J.; Bou, G.; Ceyssens, P.; Coll, P.; Corcoran, D.; Fangous, M.-S.; González-Álvarez, I.; Gorton, R.; Greub, G.; et al. How to: Identify non-tuberculous Mycobacterium species using MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2018, 24, 599–603.
- Robinne, S.; Saad, J.; Morsli, M.; Hamidou, Z.H.; Tazerart, F.; Drancourt, M.; Baron, S.A. Rapid Identification of Mycobacterium tuberculosis Complex Using Mass Spectrometry: A Proof of Concept. Front. Microbiol. 2022, 13, 753969.
- O’Connor, J.; Corcoran, G.; O’Reilly, B.; O’Mahony, J.; Lucey, B. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) for Investigation of Mycobacterium tuberculosis Complex Outbreaks: A Type Dream? J. Clin. Microbiol. 2020, 58, e02077-19.
- Kostrzewa, M.; Nagy, E.; Schröttner, P.; Pranada, A.B. How MALDI-TOF mass spectrometry can aid the diagnosis of hard-to-identify pathogenic bacteria—the rare and the unknown. Expert Rev. Mol. Diagn. 2019, 19, 667–682.
- Tsuchida, S.; Nakayama, T. MALDI-Based Mass Spectrometry in Clinical Testing: Focus on Bacterial Identification. Appl. Sci. 2022, 12, 2814.
- Lau, A.F. Matrix-assisted laser desorption ionization time-of-flight for fungal identification. Clin. Lab. Med. 2021, 41, 267–283.
- Welker, M.; Van Belkum, A.; Girard, V.; Charrier, J.-P.; Pincus, D. An update on the routine application of MALDI-TOF MS in clinical microbiology. Expert Rev. Proteom. 2019, 16, 695–710.
- Cassagne, C.; Ranque, S.; Normand, A.-C.; Fourquet, P.; Thiebault, S.; Planard, C.; Hendrickx, M.; Piarroux, R. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS ONE 2011, 6, e28425.
- Zvezdanova, M.; Escribano, P.; Ruiz, A.; Martínez-Jiménez, M.; Peláez, T.; Collazos, A.; Guinea, J.; Bouza, E.; Rodríguez-Sánchez, B. Increased species-assignment of filamentous fungi using MALDI-TOF MS coupled with a simplified sample processing and an in-house library. Med. Mycol. 2019, 57, 63–70.
- Normand, A.; Becker, P.; Gabriel, F.; Cassagne, C.; Accoceberry, I.; Gari-Toussaint, M.; Hasseine, L.; De Geyter, D.; Pierard, D.; Surmont, I. Validation of a new web application for identification of fungi by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2017, 55, 2661–2670.
- Sanguinetti, M.; Posteraro, B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2017, 55, 369–379.
- Patel, R. A moldy application of MALDI: MALDI-ToF mass spectrometry for fungal identification. J. Fungi 2019, 5, 4.
- Chen, X.-F.; Hou, X.; Xiao, M.; Zhang, L.; Cheng, J.-W.; Zhou, M.-L.; Huang, J.-J.; Zhang, J.-J.; Xu, Y.-C.; Hsueh, P.-R. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) analysis for the identification of pathogenic microorganisms: A review. Microorganisms 2021, 9, 1536.
- Sjoholm, M.I.; Dillner, J.; Carlson, J. Multiplex detection of human herpesviruses from archival specimens by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2008, 46, 540–545.
- Cai, X.; Guan, Q.; Huan, Y.; Liu, Z.; Qi, J.; Ge, S. Development of high-throughput genotyping method of all 18 HR HPV based on the MALDI-TOF MS platform and compared with the Roche Cobas 4800 HPV assay using clinical specimens. BMC Cancer 2019, 19, 825.
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front. Microbiol. 2017, 8, 1510.
- Yoshinari, T.; Hayashi, K.; Hirose, S.; Ohya, K.; Ohnishi, T.; Watanabe, M.; Taharaguchi, S.; Mekata, H.; Taniguchi, T.; Maeda, T. Matrix-Assisted Laser Desorption and Ionization Time-of-Flight Mass Spectrometry Analysis for the Direct Detection of SARS-CoV-2 in Nasopharyngeal Swabs. Anal. Chem. 2022, 94, 4218–4226.
- Thomas, J.J.; Falk, B.; Fenselau, C.; Jackman, J.; Ezzell, J. Viral characterization by direct analysis of capsid proteins. Anal. Chem. 1998, 70, 3863–3867.
- Calderaro, A.; Arcangeletti, M.C.; Rodighiero, I.; Buttrini, M.; Montecchini, S.; Simone, R.V.; Medici, M.C.; Chezzi, C.; De Conto, F. Identification of different respiratory viruses, after a cell culture step, by matrix assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Sci. Rep. 2016, 6, 36082.
- Krokhin, O.; Li, Y.; Andonov, A.; Feldmann, H.; Flick, R.; Jones, S.; Stroeher, U.; Bastien, N.; Dasuri, K.V.; Cheng, K. Mass Spectrometric Characterization of Proteins from the SARS Virus: A Preliminary Report* S. Mol. Cell. Proteom. 2003, 2, 346–356.
- Tran, N.K.; Howard, T.; Walsh, R.; Pepper, J.; Loegering, J.; Phinney, B.; Salemi, M.R.; Rashidi, H.H. Novel application of automated machine learning with MALDI-TOF-MS for rapid high-throughput screening of COVID-19: A proof of concept. Sci. Rep. 2021, 11, 8219.
- Lazari, L.C.; Zerbinati, R.M.; Rosa-Fernandes, L.; Santiago, V.F.; Rosa, K.F.; Angeli, C.B.; Schwab, G.; Palmieri, M.; Sarmento, D.J.; Marinho, C.R. MALDI-TOF mass spectrometry of saliva samples as a prognostic tool for COVID-19. J. Oral Microbiol. 2022, 14, 2043651.
- Camarasa, C.G.; Cobo, F. Application of MALDI-TOF mass spectrometry in clinical virology. In The Use of Mass Spectrometry Technology (MALDI-TOF) in Clinical Microbiology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 167–180.
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29.
- Idelevich, E.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018, 24, 738–743.
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596.
- Kerremans, J.; Verboom, P.; Stijnen, T.; Roijen, L.H.-V.; Goessens, W.; Verbrugh, H.; Vos, M. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J. Antimicrob. Chemother. 2008, 61, 428–435.
- Oviaño, M.; Bou, G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin. Microbiol. Rev. 2018, 32, e00037-18.
- Oviaño, M.; Sparbier, K.; Barba, M.J.; Kostrzewa, M.; Bou, G. Universal protocol for the rapid automated detection of carbapenem-resistant Gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS). Int. J. Antimicrob. Agents 2016, 48, 655–660.
- Hrabák, J. Detection of carbapenemases using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) meropenem hydrolysis assay. In Sepsis; Springer: Berlin/Heidelberg, Germany, 2015; pp. 91–96.
- Camara, J.E.; Hays, F.A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1633–1638.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
28 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





