Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Li, H.; Li, Z. Microbial Mass Spectrometry Imaging. Encyclopedia. Available online: https://encyclopedia.pub/entry/35964 (accessed on 07 February 2026).
Li H, Li Z. Microbial Mass Spectrometry Imaging. Encyclopedia. Available at: https://encyclopedia.pub/entry/35964. Accessed February 07, 2026.
Li, Hao, Zhiyong Li. "Microbial Mass Spectrometry Imaging" Encyclopedia, https://encyclopedia.pub/entry/35964 (accessed February 07, 2026).
Li, H., & Li, Z. (2022, November 23). Microbial Mass Spectrometry Imaging. In Encyclopedia. https://encyclopedia.pub/entry/35964
Li, Hao and Zhiyong Li. "Microbial Mass Spectrometry Imaging." Encyclopedia. Web. 23 November, 2022.
Copy Citation
As an impressive mass spectrometry technology, mass spectrometric imaging (MSI) can provide mass spectra data and spatial distribution of analytes simultaneously. MSI has been widely used in diverse fields such as clinical diagnosis, the pharmaceutical industry and environmental study due to its accuracy, high resolution and developing reproducibility. Natural products (NPs) have been a critical source of leading drugs; almost half of marketed drugs are derived from NPs or their derivatives.
mass spectrometry imaging
microorganism
natural products
1. Introduction
Bioactive microbial natural products (NPs) are important sources of leading drugs [1]. Since the discovery of penicillin [2], human exploration in the field of microbial NPs has continued for nearly 100 years. To date, microbial natural products and their derivatives have made a great contribution to human health, especially for antitumor and antimicrobial drug development [3]. Generally, in a routine microbial NPs’ research workflow, after the isolation, NPs need to be characterized by several methods for their chemical properties. As a regular analytic and characteristic technology, mass spectrometry is an indispensable tool in this field, which offers qualitative (mass-to-charge ratio) and quantitative (intensity) information for specific molecules [4][5]. However, traditional mass spectrometry analysis for microbial NPs requires laborious processes including fermentation, extraction, concentration and dissolution, which often cost days to weeks. More importantly, the compounds are removed from their original biological environment, which prevents the understanding of NPs’ real biofunctions or biosynthetic origins in site [6].
At the forefront of microbial NPs’ discovery, different kinds of advanced mass spectrometry imaging (MSI) technologies enable direct characterization and analysis of microbial NPs or microbial interaction, which allows researchers to connect the microbial phenotypic phenomena to chemical information [7]. As MSI can capture the spatial distribution of the chemical information of analytes, it combines the mass spectrum data and molecules’ spatial distribution into one experiment, which cannot be achieved by traditional mass spectrometry.
MSI initially began with solid surface analysis with secondary ion mass spectrometry (SIMS) imaging in the early 1960s, and progressed with commercial laser microprobe mass spectrometry (LMMS) [8]. In recent years, largely benefitting from the advances in the soft ionization technique, MSI has been applied rapidly in the biology field [7], since the investigation of peptides and proteins in biological samples based on matrix-assisted laser desorption/ionization (MALDI-TOF-MS) [9]. The research of microbial MSI has continued to grow in the past two decades (Figure 1). With the ability of direct untargeted investigation of diverse molecules such as glycans, lipids, peptides and macromolecules, with a label-free method on a complex biological sample surface, MSI offers a powerful tool to complement other biological imaging and omics study [10].

Figure 1. Microbial mass spectrometry imaging-related papers on Web of Science™ from 2002–2021.
As shown in Figure 2, a standard MSI workflow generally contains four steps: (1) the sample is induced to desorption under the laser or ion beam gun; (2) the gas phase is ionized for further detection in the mass analyzer (please find the detailed ionization strategy in Table 1); (3) the detector records the values of the mass-to-charge ratio (m/z) and generates a mass spectrum graph, with m/z values on the x axis and ion intensity on the y axis; and (4) the collected mass spectra of every pixel on the surface of the analytes are performed as a heat map to visualize the spatial chemical information using professional data processing software [11].
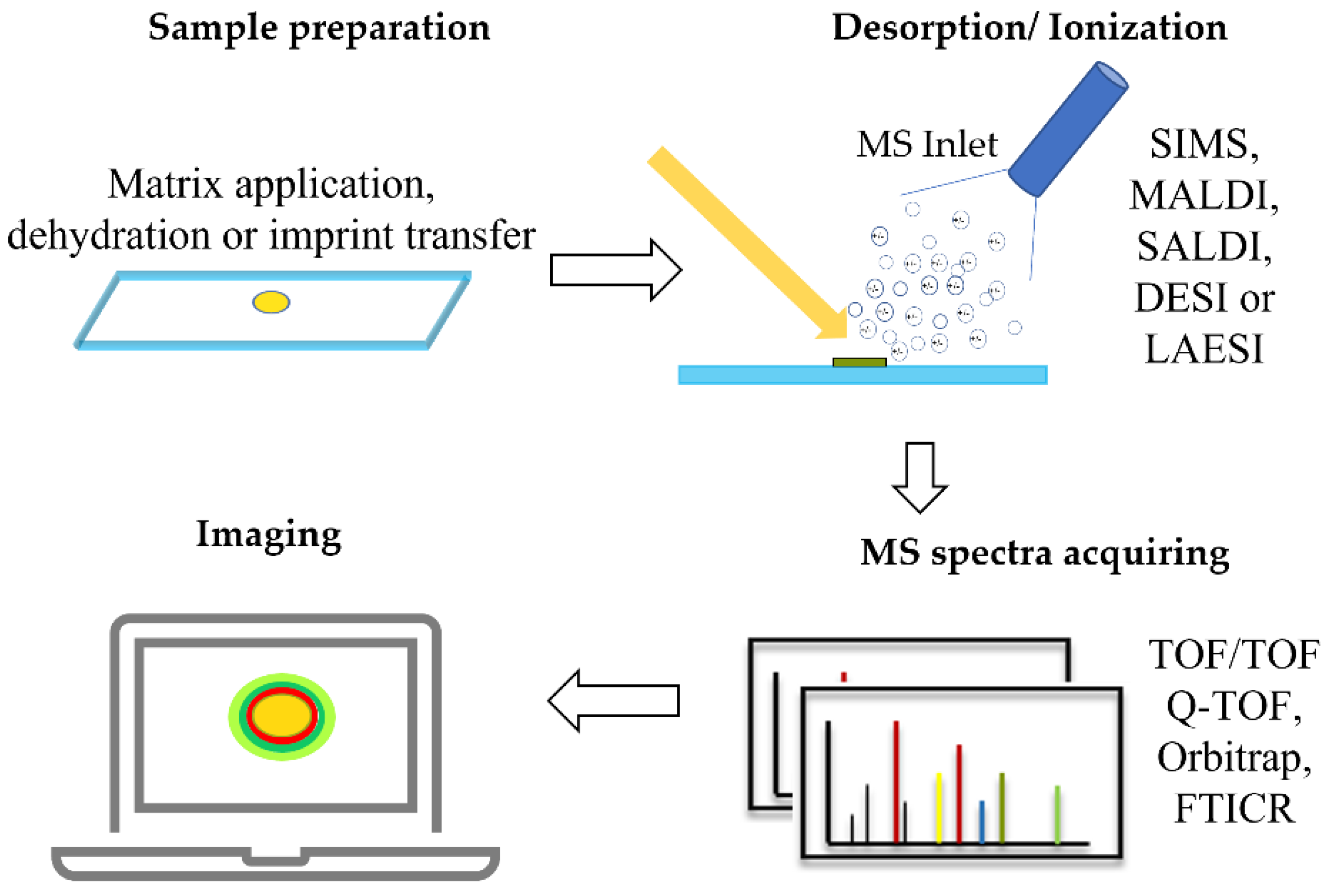
Figure 2. A typical workflow of microbial MSI. SIMS: secondary ion mass spectrometry; MALDI: matrix-assisted laser desorption/ionization; SALDI: surface-assisted laser desorption/ionization; DESI: desorption electrospray ionization; LAESI: laser ablation electrospray ionization; TOF: time-of-flight, Q-TOF: quadrupole time-of-flight, FTICR: Fourier transform ion cyclotron resonance.
Table 1. Ionization techniques used in microbial MSI.
| Ion Source Working Condition | Ionization Source | Limit of Spatial Resolution | Advantage | Major Limit |
|---|---|---|---|---|
| Vacuum ionization | SIMS | 35 nm [12] | Superior spatial resolution | Mostly fragmentation ions |
| MALDI | 5 μm [7] | Broad coverage of molecule species | The choice of matrix should be considered | |
| SALDI | 10 μm [7] | Matrix-free | Sample transfer is inevitable | |
| Ambient ionization | DESI | 10 μm [13][14] | Simple sample preparation | Complex instrumental parameters optimization |
| LAESI | 100 μm [15] | Can be used for fresh samples | Lower spatial resolution |
It is worth noting that the application of MSI in microbial NPs’ research has led to great achievements [7][11][16][17][18][19][20][21][22][23][24][25][26].
2. Different Ionization Used in Microbial MSI
For any mass spectrometry analysis, the ionization level and efficiency of the analytes should be considered as one of the most important factors [7][27]. To date, diverse commercial ion sources have been used for microbial MSI (Table 1). Generally, researchers divide them into two sections—“vacuum ionization” and “ambient ionization”—according to the ion source working condition [17]. Secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption/ionization (MALDI), surface-assisted laser desorption/ionization (SALDI), desorption electrospray ionization (DESI) and laser ablation electrospray ionization (LAESI) are the five major MSI methods in the field of microbial NPs. Spraker et al. recently reviewed the ionizations in MSI for NPs’ research that can be used as a guide for those seeking further background reading [7].
2.1. Secondary Ion Mass Spectrometry (SIMS)
As the earliest MSI technology used for microbial research [28], SIMS depends on the charge transfer from the high-energy primary ion beam to the sample surface for generating the analyte secondary ions. SIMS can provide superior spatial resolution (<100 nm), far beyond other MSI technologies [29]. However, the harsh requirements for the surface of samples limit the application of SIMS in the exploration of microbial NPs. Cultivating microbial samples on conductive substrate surfaces such as silicon wafer and indium tin oxide-coated (ITO) glass slides can partly solve this problem [30][31]. On the other hand, due to the nature of the hard ionization technique, SIMS produces some poorly interpretable secondary fragmentation ions, which limits the application in the identification of unknown natural products [11]. As a means of optimization, matrix-enhanced SIMS (ME-SIMS) can partly help to reduce fragmentation, enhance ionization and extend the mass range of SIMS experiments [32].
2.2. Matrix-Assisted Laser Desorption/Ionization (MALDI)
In 1985, MALDI was firstly discovered by Franz Hillencamp and Michael Karas [33], and then Koichi Tanaka developed this technology into protein analysis and won the Nobel Prize in 2002 [34]. Due to the broad coverage of molecule species (small molecules, lipopeptides, peptides and proteins), MALDI has become the first choice of microbial MSI [35]. In MALDI-MSI, biomolecules on the sample surface are desorbed by the assistance of laser light and the matrix (small organic acids or bases that cocrystallize with the sample) for proper ionization [36]. Hence, the selection of a suitable matrix is considered as one of the critical factors, especially for NP research [37]. Until now, 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA) and their mixture have been the most common choices for various small molecule imaging in positive ion mode [38], while in negative ion mode, 9-aminoacridine (9-AA) has shown the advantage of low background with MALDI-MSI [39]. In addition, 3,5-dimethoxy-4-hydroxycinnamic acid (SA, sinapinic acid) is used for protein analysis, and 4,6-Trihydroxyacetophenone (THAP) [40] and 3-Hydroxypicolinic acid (3-HPA) [41] are justified to be suitable for oligonucleotide analysis. In addition, many other matrices substrates such as Dithranol [42], Curcumin [43], 4-Phenly-α-cyanocinnamic acid amide [44], 1,6-diphenyl-1,3,5-hexatriene (DPH) [45], 3-Aminophthalhydrazide (Luminol) [38], N-phenyl-2-naphthylamine (PNA) [46], 1,5-diaminonapthalene (1,5-DAN) [47], 1-naphthylhydrazine hydrochloride (NHHC) [48] have been used. Meanwhile scientists are still searching for new matrices with better performance.
2.3. Surface-Assisted Laser Desorption/Ionization (SALDI)
SALDI-MS was originally proposed by Sunner and Chen in 1995 [49]. As a kind of matrix-free laser desorption/ionization (LDI) technology, SALDI substitutes the organic matrix of MALDI with other substrate surfaces such as graphite or nano silicon [7]. Due to the avoidance of organic matrices, SALDI provides the ideal prospect for application in small molecules (<500 Da) [50][51]. In the past decade, there were several applications of biological and microbial SALDI imaging based on different kinds of silicon substrates [52][53][54]. For instance, Wang et al. prepared gold nanoparticles/thiol-β-cyclodextrin-functionalized TiO2 nanowires as the assistant surface for NP SALDI-MSI in 2022 [50]. Meanwhile, several studies have indicated that SALDI could be a promising platform for microbial imaging [25][55].
2.4. Desorption Electrospray Ionization (DESI)
In 2004, as a novel ambient ionization technology, DESI was firstly innovated by Cooks’ group [56]. In a standard DESI process, the high-velocity electrosolvent is directly sprayed on the surface of the sample to form sample-bearing droplets, and then the ions generated by desolvation of these droplets are introduced to the mass spectrometer inlet [18]. Routinely, in order to obtain ideal imaging results, the optimization for instrument parameters is inevitable, such as solvent selection, solvent flow rate, spray voltage, gas flow rate, and the distances and angle from the sample surface to the sprayer [11]. However, DESI-MSI requires a hard, flat, uniform and nonconductive sample surface for the consistent and stable signal, which partly limits the application of DESI-MSI to microbial NP analysis [57].
2.5. Laser Ablation Electrospray Ionization (LAESI)
LAESI was invented in 2007 by Nemes and Vertes [58] and provided a new choice for ambient ionization mass spectrometry technology via the combination of electrospray ionization and laser ablation. Gas particles on the analyte surface, formed by mid-infrared laser ablation, are introduced to the MS inlet for analysis after electrospray ionization. LAESI requires little sample preparation but samples should be water-rich, which allows direct analysis of some limited sample surfaces such as plant tissues, clinical samples and microorganisms [59]. In 2016, Li et al. first reported the application of LAESI-MSI in a bacterial colony on agar to show the bacterial lipids’ distribution, which presented its ability in the identification and imagination of microbial metabolites [60].
3. Sample Preparation in Microbial MSI
Unlike usual vegetal or mammalian tissue section imaging, sample preparation for microbial samples in MSI is a tricky problem [25]. Flat frozen sections of organs or tissues can be easily obtained from cryogenic microtomes, but, unfortunately, microbial agar cultures are often too thin for cryotome sectioning. In addition, most microbial samples contain complex topography due to the presence of spores and aerial hyphae on the hydrated agar media, which makes it difficult to meet the requirements of the ion source for the analyte surface. For those vacuum ion sources such as MALDI, it demands a dry sample and homogeneous deposition of the specific matrix. Though it is a tough challenge, much useful and impressive progress has been made this decade, since the first protocol for agar-based microbial MALDI-MSI was introduced [61]. A typical microbial sample preparation contains three steps: (1) culturing the microbe on thin agar; (2) dehydrating the sample; and (3) depositing the matrix on the sample surface. Actually, the factors to be considered in each specific experimental process are far more complicated than the three steps above. There are many details to be optimized such as the agar concentration of the culture media, the choice of matrix and the matrix deposition methods (including sieving, sublimation, using an airbrush and a robotic sprayer) [25]. Some advances in the sample preparation of microbial MALDI-MSI are summarized in Table 2. It should be noticed that, due to the construction of the instrument and unstable sample adherence, the agar sample placed on the MALDI target plate may flake or even fall from the plate under a high-vacuum condition, which brings disadvantages to the experiment and even causes irreversible damage to the instrument. Cultivating microbes on a glass slide coated with indium tin oxide (ITO) or some optimization of the sample preparation can partly overcome this problem [35][61]. Meanwhile, AP-MALDI was subsequently developed and has been performed to monitor the spatial metabolome of the host–microbe associations of deep-sea mussel Bathymodiolus puteoserpentis at atmospheric pressure, which will alleviate this vacuum restriction problem [10][62].
Table 2. Technical advances in sample preparation of microbial MALDI-MSI.
| Time | Introduced Technique | Advantage | Reference |
|---|---|---|---|
| 2012 | A common protocol for agar-based microbial MALDI-MSI | The first protocol for agar-based microbial MALDI-MSI | [61] |
| 2013 | A method for visible three-dimensional (3D) models of a microbial colony | Captures the depth profile of metabolite distribution beyond 2D-MALDI-MSI | [63] |
| 2014 | A method with solid MALDI matrix deposition on microbial agar culture | Enhanced signals of some fungal metabolites | [64] |
| 2015 | A method for spraying matrix solution programmatically on dried agar-based samples | Forms a homogeneous, evenly closed matrix layer | [65] |
| 2016 | A robotic matrix sprayer with a heated capillary | Higher sensitivity and lateral resolution for the analyte and suitable for different kinds of matrix | [66] |
| 2016 | A one-step matrix spraying method with optimized homemade equipment | Simplified sample preparation and high-resolved images | [67] |
| 2019 | A membrane-based culturing workflow | Offers a safe and flat microbial sample surface | [68] |
| 2022 | A method using 2,5-dihydroxybenzoic acid (DHB) as “glue” to adhere the microbial culture agar plate to the MALDI target | Prevents the sample flaking from the target under vacuum and also provides a larger area for MALDI-MSI analysis | [35] |
DESI-MSI, as the other mainstream option used for NP or microbial metabolite imaging, requires relatively simple sample preparation compared to MALDI-MSI. Some technical advances in the sample preparation of microbial DESI-MSI are summarized in Table 3. Routine DESI-MSI analysis demands a hard, flat and uniform surface. Several viable alternative solutions have been developed, for example, imprint transfer technology, the involved metabolites and its distribution on the surface of the microbial culture medium is transferred to mixed cellulose ester filter membranes, and thus DESI analysis can be performed on this flat membrane [57]. However, this method might lose some molecular information and is not ideal for those microbial samples with complex surface conditions such as fungi [69]. Another simple but exciting advance in microbial sample preparation for DESI-MSI is dehydration. For microbes grown on a thin-layered agar plate, the extra dehydration can form a flat, hard and nonconductive surface, which is suitable for direct DESI-MSI analysis [69]. Nano-DESI-MSI technology has the ability of minimally invasive and direct analysis of a living microbial community, without any sample preparation [70].
Table 3. Technical advances in sample preparation of microbial DESI-MSI.
| Time | Introduced Technique | Advantage | Reference |
|---|---|---|---|
| 2010 | A thin film imprinting technique with mixed cellulose ester filter membranes | Uses a complementary surface to make an imprint of the bacterial culture from solid agar for imaging | [57] |
| 2012 | Nano-DESI-MSI technology | Direct chemical monitoring of living microbial colonies grown on a Petri dish | [70] |
| 2014 | A “cardboard insert” method | An effective method for fungal culture imaging in situ with a hard and flat surface | [71] |
| 2015 | A protocol on microbial agar culture for direct DESI-MSI | Offers rapid sample preparation with a dehydrated and hard surface for imaging | [69] |
| 2017 | A constant-distance nano-DESI-MSI imaging model | An ideal method for imaging microbial samples with complex topography | [72] |
| 2019 | A microbial sample reparation method with microporous membrane scaffolds (MMS) | An effective method for imaging and evaluating microbial interspecies interactions | [73] |
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803.
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. Influenzae Br. J. Exp. Pathol. 1929, 10, 226.
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404.
- Aksenov, A.A.; da Silva, R.; Knight, R.; Lopes, N.P.; Dorrestein, P.C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 1–20.
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug. Discov. 2021, 20, 200–216.
- Simmons, T.L.; Coates, R.C.; Clark, B.R.; Engene, N.; Gonzalez, D.; Esquenazi, E.; Dorrestein, P.C.; Gerwick, W.H. Biosynthetic origin of natural products isolated from marine microorganism–invertebrate assemblages. Proc. Natl. Acad. Sci. USA 2008, 105, 4587–4594.
- Spraker, J.E.; Luu, G.T.; Sanchez, L.M. Imaging mass spectrometry for natural products discovery: A review of ionization methods. Nat. Prod. Rep. 2020, 37, 150–162.
- Amstalden van Hove, E.R.; Smith, D.F.; Heeren, R.M. A concise review of mass spectrometry imaging. J. Chromatogr. A. 2010, 1217, 3946–3954.
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760.
- Geier, B.; Sogin, E.M.; Michellod, D.; Janda, M.; Kompauer, M.; Spengler, B.; Dubilier, N.; Liebeke, M. Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nat. Microbiol. 2020, 5, 498–510.
- Esquenazi, E.; Yang, Y.-L.; Watrous, J.; Gerwick, W.H.; Dorrestein, P.C. Imaging mass spectrometry of natural products. Nat. Prod. Rep. 2009, 26, 1521–1534.
- Fletcher, J.S.; Vickerman, J.C. Secondary ion mass spectrometry: Characterizing complex samples in two and three dimensions. Anal. Chem. 2013, 85, 610–639.
- He, J.; Luo, Z.; Huang, L.; He, J.; Chen, Y.; Rong, X.; Jia, S.; Tang, F.; Wang, X.; Zhang, R.; et al. Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Anal. Chem. 2015, 87, 5372–5379.
- Yin, R.; Burnum-Johnson, K.E.; Sun, X.; Dey, S.K.; Laskin, J. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat. Protoc. 2019, 14, 3445–3470.
- Kulkarni, P.; Wilschut, R.A.; Verhoeven, K.J.F.; van der Putten, W.H.; Garbeva, P. LAESI mass spectrometry imaging as a tool to differentiate the root metabolome of native and range-expanding plant species. Planta 2018, 248, 1515–1523.
- Shih, C.J.; Chen, P.Y.; Liaw, C.C.; Lai, Y.M.; Yang, Y.L. Bringing microbial interactions to light using imaging mass spectrometry. Nat. Prod. Rep. 2014, 31, 739–755.
- Ho, Y.N.; Shu, L.J.; Yang, Y.L. Imaging mass spectrometry for metabolites: Technical progress, multimodal imaging, and biological interactions. Wiley. Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1387.
- Parrot, D.; Papazian, S.; Foil, D.; Tasdemir, D. Imaging the unimaginable: Desorption electrospray ionization—Imaging mass spectrometry (DESI-IMS) in natural product research. Planta. Med. 2018, 84, 584–593.
- Dunham, S.J.; Ellis, J.F.; Li, B.; Sweedler, J.V. Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res. 2017, 50, 96–104.
- Sgobba, E.; Daguerre, Y.; Giampa, M. Unravel the local complexity of biological environments by MALDI mass spectrometry imaging. Int. J. Mol. Sci. 2021, 22, 12393.
- Stasulli, N.M.; Shank, E.A. Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. FEMS. Microbiol. Rev. 2016, 40, 807–813.
- Fang, J.; Dorrestein, P.C. Emerging mass spectrometry techniques for the direct analysis of microbial colonies. Curr. Opin. Microbiol. 2014, 19, 120–129.
- Yang, H.; Goodlett, D.R.; Ernst, R.K.; Scott, A.J. Mass spectrometry imaging of microbes. Mass. Spectrom. Lett. 2020, 11, 41–51.
- Perez, C.J.; Bagga, A.K.; Prova, S.S.; Yousefi Taemeh, M.; Ifa, D.R. Review and perspectives on the applications of mass spectrometry imaging under ambient conditions. Rapid. Commun. Mass. Spectrom. 2019, 33, 27–53.
- Zou, Y.; Tang, W.; Li, B. Mass spectrometry imaging and its potential in food microbiology. Int. J. Food. Microbiol. 2022, 371, 109675.
- Xue, J.; Bai, Y.; Liu, H. Recent advances in ambient mass spectrometry imaging. Trends. Anal. Chem. 2019, 120, 115659.
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2012, 9, 44–66.
- Cliff, J.B.; Gaspar, D.J.; Bottomley, P.J.; Myrold, D.D. Exploration of inorganic C and N assimilation by soil microbes with time-of-flight secondary ion mass spectrometry. Appl. Environ. Microbiol. 2002, 68, 4067–4073.
- Kollmer, F.; Paul, W.; Krehl, M.; Niehuis, E. Ultra high spatial resolution SIMS with cluster ions—Approaching the physical limits. Surf. Interface. Anal. 2013, 45, 312–314.
- Lanni, E.J.; Masyuko, R.N.; Driscoll, C.M.; Dunham, S.J.; Shrout, J.D.; Bohn, P.W.; Sweedler, J.V. Correlated imaging with C60-SIMS and confocal Raman microscopy: Visualization of cell-scale molecular distributions in bacterial biofilms. Anal. Chem. 2014, 86, 10885–10891.
- Davies, S.K.; Fearn, S.; Allsopp, L.P.; Harrison, F.; Ware, E.; Diggle, S.P.; Filloux, A.; McPhail, D.S.; Bundy, J.G. Visualizing antimicrobials in bacterial biofilms: Three-dimensional biochemical imaging using TOF-SIMS. mSphere 2017, 2, e00211–e00217.
- Watrous, J.D.; Dorrestein, P.C. Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 2011, 9, 683–694.
- Karas, M.; Bachmann, D.; Hillenkamp, F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem. 1985, 57, 2935–2939.
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153.
- Jens, J.N.; Breiner, D.J.; Phelan, V.V. Spray-based application of matrix to agar-based microbial samples for reproducible sample adherence in MALDI MSI. J. Am. Soc. Mass. Spectrom. 2022, 33, 731–734.
- Silva, R.; Lopes, N.P.; Silva, D.B. Application of MALDI mass spectrometry in natural products analysis. Planta. Med. 2016, 82, 671–689.
- Vickerman, J.C. Molecular imaging and depth profiling by mass spectrometry—SIMS, MALDI or DESI? Analyst 2011, 136, 2199–2217.
- Li, B.; Sun, R.; Gordon, A.; Ge, J.; Zhang, Y.; Li, P.; Yang, H. 3-Aminophthalhydrazide (Luminol) as a matrix for dual-polarity MALDI MS imaging. Anal. Chem. 2019, 91, 8221–8228.
- Edwards, J.L.; Kennedy, R.T. Metabolomic analysis of eukaryotic tissue and prokaryotes using negative mode MALDI time-of-flight mass spectrometry. Anal. Chem. 2005, 77, 2201–2209.
- Tholey, A.; Heinzle, E. Ionic (liquid) matrices for matrix-assisted laser desorption/ionization mass spectrometry—Applications and perspectives. Anal. Bioanal. Chem. 2006, 386, 24–37.
- Pan, C.; Xu, S.; Zhou, H.; Fu, Y.; Ye, M.; Zou, H. Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Anal. Bioanal. Chem. 2007, 387, 193–204.
- Le, C.H.; Han, J.; Borchers, C.H. Dithranol as a MALDI matrix for tissue imaging of lipids by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2012, 84, 8391–8398.
- Francese, S.; Bradshaw, R.; Flinders, B.; Mitchell, C.; Bleay, S.; Cicero, L.; Clench, M. Curcumin: A multipurpose matrix for MALDI mass spectrometry imaging applications. Anal. Chem. 2013, 85, 5240–5248.
- Fülöp, A.; Porada, M.B.; Marsching, C.; Blott, H.; Meyer, B.; Tambe, S.; Sandhoff, R.; Junker, H.-D.; Hopf, C. 4-Phenyl-α-cyanocinnamic acid amide: Screening for a negative ion matrix for MALDI-MS imaging of multiple lipid classes. Anal. Chem. 2013, 85, 9156–9163.
- Ibrahim, H.; Jurcic, K.; Wang, J.S.-H.; Whitehead, S.N.; Yeung, K.K.-C. 1, 6-Diphenyl-1, 3, 5-hexatriene (DPH) as a novel matrix for MALDI MS imaging of fatty acids, phospholipids, and sulfatides in brain tissues. Anal. Chem. 2017, 89, 12828–12836.
- Liu, H.; Zhou, Y.; Wang, J.; Xiong, C.; Xue, J.; Zhan, L.; Nie, Z. N-Phenyl-2-naphthylamine as a novel MALDI matrix for analysis and in situ imaging of small molecules. Anal. Chem. 2018, 90, 729–736.
- Thomas, A.l.; Charbonneau, J.L.; Fournaise, E.; Chaurand, P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: Enhanced information in both positive and negative polarities after 1, 5-diaminonapthalene deposition. Anal. Chem. 2012, 84, 2048–2054.
- He, Q.; Chen, S.; Wang, J.; Hou, J.; Wang, J.; Xiong, S.; Nie, Z. 1-Naphthylhydrazine hydrochloride: A new matrix for the quantification of glucose and homogentisic acid in real samples by MALDI-TOF MS. Clin. Chim. Acta. 2013, 420, 94–98.
- Law, K.; Larkin, J.R. Recent advances in SALDI-MS techniques and their chemical and bioanalytical applications. Anal. Bioanal. Chem. 2011, 399, 2597–2622.
- Wang, X.N.; Li, B. Monolithic gold nanoparticles/thiol-beta-cyclodextrin-functionalized TiO2 nanowires for enhanced SALDI MS detection and imaging of natural products. Anal. Chem. 2022, 94, 952–959.
- Song, K.; Cheng, Q. Desorption and ionization mechanisms and signal enhancement in surface assisted laser desorption ionization mass spectrometry (SALDI-MS). Appl. Spectrosc. Rev. 2019, 55, 220–242.
- Ronci, M.; Rudd, D.; Guinan, T.; Benkendorff, K.; Voelcker, N.H. Mass spectrometry imaging on porous silicon: Investigating the distribution of bioactives in marine mollusc tissues. Anal. Chem. 2012, 84, 8996–9001.
- Louie, K.B.; Bowen, B.P.; Cheng, X.; Berleman, J.E.; Chakraborty, R.; Deutschbauer, A.; Arkin, A.; Northen, T.R. “Replica-extraction-transfer” nanostructure-initiator mass spectrometry imaging of acoustically printed bacteria. Anal. Chem. 2013, 85, 10856–10862.
- Chen, P.Y.; Hsieh, C.Y.; Shih, C.J.; Lin, Y.J.; Tsao, C.W.; Yang, Y.L. Exploration of fungal metabolic interactions using imaging mass spectrometry on nanostructured silicon. J. Nat. Prod. 2018, 81, 1527–1533.
- Muller, W.H.; Verdin, A.; De Pauw, E.; Malherbe, C.; Eppe, G. Surface-assisted laser desorption/ionization mass spectrometry imaging: A review. Mass. Spectrom. Rev. 2022, 41, 373–420.
- Takats, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473.
- Watrous, J.; Hendricks, N.; Meehan, M.; Dorrestein, P.C. Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal. Chem. 2010, 82, 1598–1600.
- Nemes, P.; Vertes, A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem. 2007, 79, 8098–8106.
- Nemes, P.; Barton, A.A.; Li, Y.; Vertes, A. Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization mass spectrometry. Anal. Chem. 2008, 80, 4575–4582.
- Li, H.; Balan, P.; Vertes, A. Molecular imaging of growth, metabolism, and antibiotic inhibition in bacterial colonies by laser ablation electrospray ionization mass spectrometry. Angew. Chem. Int. Ed. Engl. 2016, 55, 15035–15039.
- Yang, J.Y.; Phelan, V.V.; Simkovsky, R.; Watrous, J.D.; Trial, R.M.; Fleming, T.C.; Wenter, R.; Moore, B.S.; Golden, S.S.; Pogliano, K.; et al. Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 2012, 194, 6023–6028.
- Qin, L.; Zhang, Y.; Liu, Y.; He, H.; Han, M.; Li, Y.; Zeng, M.; Wang, X. Recent advances in matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI-MSI) for in situ analysis of endogenous molecules in plants. Phytochem. Anal. 2018, 29, 351–364.
- Watrous, J.D.; Phelan, V.V.; Hsu, C.-C.; Moree, W.J.; Duggan, B.M.; Alexandrov, T.; Dorrestein, P.C. Microbial metabolic exchange in 3D. ISME J. 2013, 7, 770–780.
- Vergeiner, S.; Schafferer, L.; Haas, H.; Muller, T. Improved MALDI-TOF microbial mass spectrometry imaging by application of a dispersed solid matrix. J. Am. Soc. Mass. Spectrom. 2014, 25, 1498–1501.
- Hoffmann, T.; Dorrestein, P.C. Homogeneous matrix deposition on dried agar for MALDI imaging mass spectrometry of microbial cultures. J. Am. Soc. Mass. Spectrom. 2015, 26, 1959–1962.
- Anderton, C.R.; Chu, R.K.; Tolic, N.; Creissen, A.; Pasa-Tolic, L. Utilizing a robotic sprayer for high lateral and mass resolution MALDI FT-ICR MSI of microbial cultures. J. Am. Soc. Mass. Spectrom. 2016, 27, 556–559.
- Li, B.; Comi, T.J.; Si, T.; Dunham, S.J.; Sweedler, J.V. A one-step matrix application method for MALDI mass spectrometry imaging of bacterial colony biofilms. J. Mass. Spectrom. 2016, 51, 1030–1035.
- Brockmann, E.U.; Steil, D.; Bauwens, A.; Soltwisch, J.; Dreisewerd, K. Advanced methods for MALDI-MS imaging of the chemical communication in microbial communities. Anal. Chem. 2019, 91, 15081–15089.
- Angolini, C.F.; Vendramini, P.H.; Araujo, F.D.; Araujo, W.L.; Augusti, R.; Eberlin, M.N.; de Oliveira, L.G. Direct protocol for ambient mass spectrometry imaging on agar culture. Anal. Chem. 2015, 87, 6925–6930.
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752.
- Sica, V.P.; Raja, H.A.; El-Elimat, T.; Oberlies, N.H. Mass spectrometry imaging of secondary metabolites directly on fungal cultures. RSC Adv. 2014, 4, 63221–63227.
- Nguyen, S.N.; Liyu, A.V.; Chu, R.K.; Anderton, C.R.; Laskin, J. Constant-distance mode nanospray desorption electrospray ionization mass spectrometry imaging of biological samples with complex topography. Anal. Chem 2017, 89, 1131–1137.
- Ellis, B.M.; Fischer, C.N.; Martin, L.B.; Bachmann, B.O.; McLean, J.A. Spatiochemically profiling microbial interactions with membrane scaffolded desorption electrospray ionization-ion mobility-imaging mass spectrometry and unsupervised segmentation. Anal. Chem. 2019, 91, 13703–13711.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
23 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




