Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ali Movahedi | -- | 4551 | 2022-11-22 01:47:02 | | | |
| 2 | Rita Xu | Meta information modification | 4551 | 2022-11-22 04:30:21 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Movahedi, A.; Aghaei-Dargiri, S.; Barati, B.; Kadkhodaei, S.; Wei, H.; Sangari, S.; Yang, L.; Xu, C. Genetic Effects on Plant Immunity. Encyclopedia. Available online: https://encyclopedia.pub/entry/35607 (accessed on 08 February 2026).
Movahedi A, Aghaei-Dargiri S, Barati B, Kadkhodaei S, Wei H, Sangari S, et al. Genetic Effects on Plant Immunity. Encyclopedia. Available at: https://encyclopedia.pub/entry/35607. Accessed February 08, 2026.
Movahedi, Ali, Soheila Aghaei-Dargiri, Bahram Barati, Saeid Kadkhodaei, Hui Wei, Sirous Sangari, Liming Yang, Chen Xu. "Genetic Effects on Plant Immunity" Encyclopedia, https://encyclopedia.pub/entry/35607 (accessed February 08, 2026).
Movahedi, A., Aghaei-Dargiri, S., Barati, B., Kadkhodaei, S., Wei, H., Sangari, S., Yang, L., & Xu, C. (2022, November 22). Genetic Effects on Plant Immunity. In Encyclopedia. https://encyclopedia.pub/entry/35607
Movahedi, Ali, et al. "Genetic Effects on Plant Immunity." Encyclopedia. Web. 22 November, 2022.
Copy Citation
An immune system is a protective mechanism that shields plants from environmental stresses. This primary function is to maintain optimal circumstances for the growth and development of plant tissues while avoiding harm from biotic and abiotic stress factors. Plants subjected to various stressors initiate stress signaling cascades that affect multiple gene expressions and induce adaptation. These signaling pathways are coordinated by transcription factors, non-coding RNAs, RNA-binding proteins, and protein-protein interaction networks.
plant immunity
signal transduction
histone modification
1. Introduction
Undoubtedly, all living organisms' survival depends on plants. Most food, sanitary, and industrial products are derived directly or indirectly from plants. Given the high conservation of different organisms in biological systems, plant discoveries also have important implications for human health. According to the abovementioned, the importance of plants' survival and development becomes more apparent. Plant immunity is the innate or induced capacity of plants to eliminate pathogens such as bacteria, fungi, viruses, nematodes, and insects. Plants have also developed immune mechanisms to avoid herbivores [1]. Plant defense against abiotic or biotic perturbations is critical for plant adaptability and survival under suboptimal growth or adverse conditions.
Despite the many similarities between plant and animal immunities, there are also notable differences. For instance, plants lack antibodies, T cells, and circulating immune cells [2]. Plant immune responses can be categorized into functionally different classes: (1) basal responses, including the transcriptional regulation of genes in response to pathogen-associated molecular patterns (PAMPs), (2) hypersensitive responses, which involve the apoptosis of cells at the site of infection, (3) systemic acquired protection, which renders the entire plant resistant to infection, (4) jasmonic acid (JA)/ethylene (ET) pathway responses, which protect the whole plant and neighboring plants from herbivores, and (5) non-host immunity [3][4][5][6].
Environmental changes are usually unfavorable, posing severe threats to plant growth and development [7]. Abiotic stresses under adverse ecological conditions include drought, high or low temperatures, lack of nutrients, excess salt, and heavy metals in the soil. Besides biotic stresses, abiotic stresses significantly affect crop yield and wild plant species distribution. Drought, salinity, and extreme temperatures are the abiotic stresses affecting the geographic distribution and limiting crop yield. The harmful effects of biological and abiotic stresses on plants are worsening because extreme weather is occurring more often [8]. In a natural environment, the ability of plants to cope with abiotic stresses is influenced by microbial communities composed of bacteria, fungi, and oomycetes that inhabit the host plant without causing disease. Upon recognizing environmental stresses via specific sensing mechanisms, plants adapt their growth, metabolism, and development. Hence, like almost all living organisms, plants have developed signaling pathways to sense changes in their environment and adjust their metabolism and biological functions to prevent stress-related damage [7]. Osmotic stress caused by drought has been recognized as the most common abiotic stress, followed by salinity stress [9], causing ion toxicity [7]. The secondary effects of drought and salinity stress are incredibly complex. They include oxidative stress, metabolic dysfunction, and damage to various cell components, such as membrane lipids, proteins, and nucleic acids [10]. Although primary stress signals can induce immune responses in plants, secondary stress signals are the main triggers of plant immune responses [11]. Extensive crosstalk occurs between the signal transduction pathways induced by drought and salt stress. For instance, the hyperosmotic signals induced by drought and salt stress promote the accumulation of the plant hormone abscisic acid (ABA), which is responsible for numerous plant biological responses [12].
2. Biological Defenses Mediate Plant Immunity
The crosstalk among hormone signal transduction pathways in response to invading pathogens can be antagonistic or synergistic. Salicylic acid (SA) and JA are the predominant hormonal pathways in plant defense [13]. Plant defense mechanisms are versatile and tailored according to the pathogens' characteristics. The SA pathway is induced primarily in response to biotrophic pathogens, whereas the JA/ET pathway is involved in the defense against herbivore or necrotrophic pathogen infections [14]. Typically, only one of these pathways is activated to preserve energy and resources [15]. Although JA, ET, and SA are the predominant hormones in plant defense, additional hormones play a supporting role. These hormones include ABA, auxins, gibberellins, cytokinins, and brassinosteroids. SA has a vital role in the fight against biotrophic pathogens, whereas JA and ET are critical defense hormones in response to herbivores and necrotrophic pathogens. While the JA/ET and SA pathways have been the most antagonistic, studies in mutant plants have suggested a synergistic effect under certain conditions [16]. In pattern-triggered immunity (PTI), PAMPs such as bacterial flagellin and fungal chitin are detected by transmembrane pattern recognition receptors (PRRs), which initiate PTI on the cell surface. Upon activation, PRRs initiate complex defense signaling cascades [17].
Plant immunity relies on innate immune receptors, which are expressed in every cell and recognize invasion signals, inducing PTI or ETI [18]. To prevent pathogen infection, plants use receptor barriers for innate defense. Microbial or altered host molecules are recognized by PRRs in the plasma membrane, activating PTI [19][20]. Damaged endogenous molecules, microbe-associated molecular patterns (MAMPs), and danger-associated molecular patterns are recognized and bound to by PRRs. Activation of PRR by cytosolic Ca2+ and apoplastic reactive oxygen species (ROS) responses promotes phosphorylation and activates the NADPH oxidase respiratory burst oxidase D (RBOHD). RBOHD activation results in the rapid production of ROS in a calcium-dependent or calcium-independent manner. Increases in ROS and cytosolic Ca2+ cause the activation of Ca2+-dependent protein kinases, mitogen-activated protein kinases (MAPKs), and defense hormone networks, as well as a lot of transcriptional, translational, and metabolic reprogramming [21] (Figure 1).
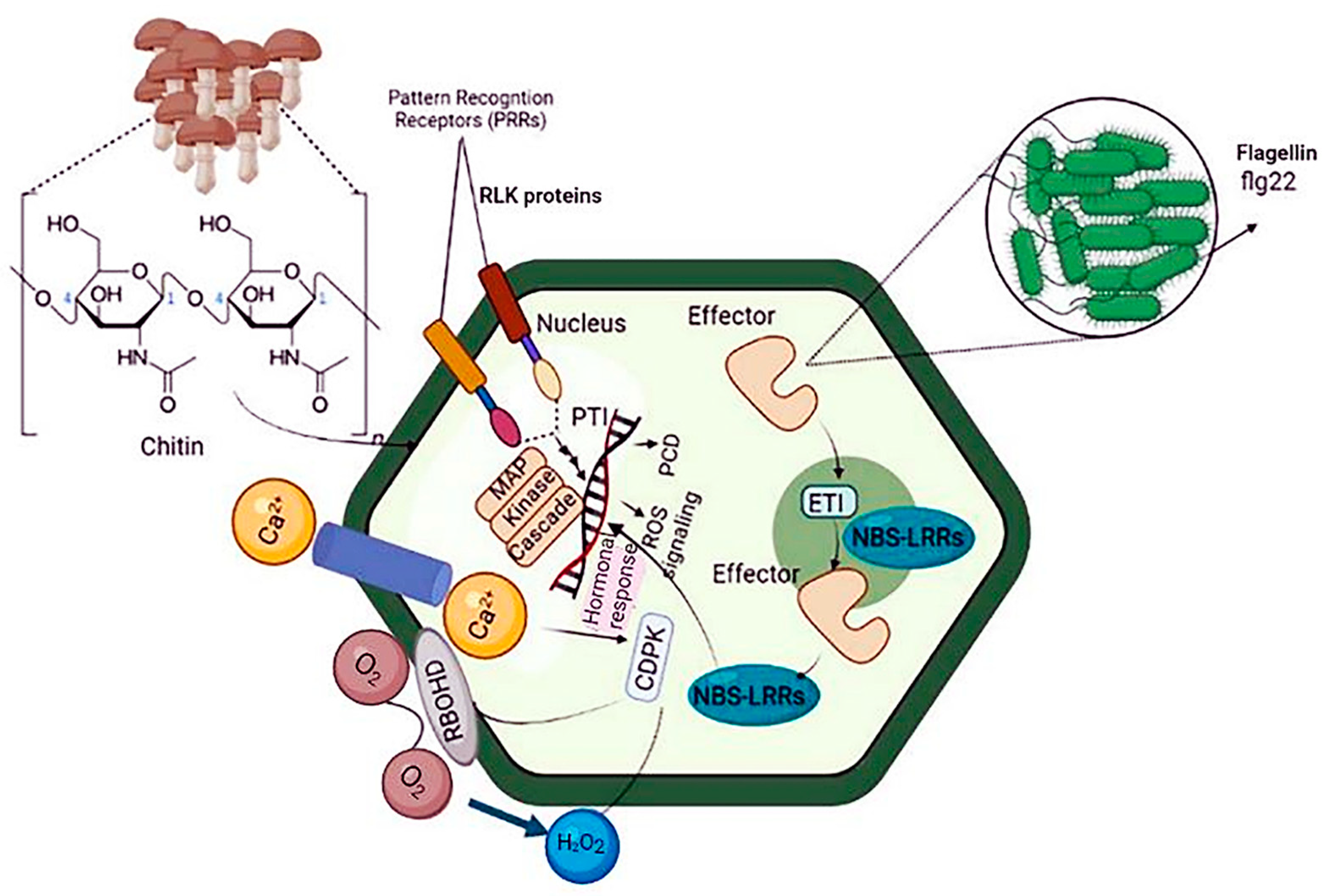
Figure 1. Schematic diagram of plant immunity against bacterial pathogens. Plants use two strategies to respond to pathogen attacks: PTI and ETI. Because ETI recognizes specific proteins injected by the pathogen rather than PAMPs, ubiquitous molecules expressed by all microorganisms, it is more pathogen strain-specific than PTI. The plant must express a resistance protein that recognizes a specific factor protein for ETI to occur. PTI: pattern-triggered immunity, ETI: effector-triggered immunity, and PAMP: pathogen-associated molecular patterns.
MicroRNAs (miRNAs) are central players in these defense cascades, modulating gene expression, metabolism, and plant development [22]. PTI results in distinct physiological phenomena, including stomata closure to prevent pathogen penetration; ROS and nitric oxide (NO) production; translocation of nutrients from the cytosol to the apoplast; deposition of callose; and production of various antimicrobial metabolites and defense hormones [23][24]. Signaling cascades involved in ETI are more robust than PTI-related cascades. ETI is initiated upon plant recognition of pathogen virulence factors known as effector molecules by resistance proteins (R proteins). R proteins can be extracellular or intracellular and recognize pathogen virulence factors directly or indirectly. Most R proteins contain nucleotide-binding sites and leucine-rich repeat domains. The gene family containing these domains is conserved across plant species and includes highly polymorphic genes often found in clusters [25]. Thus, PTI and ETI result in extensive reprogramming of plant gene expression via various receptor proteins, signal transduction cascades, kinases, ROS, hormones, heat shock proteins, and transcription factors, protecting against invading pathogens [26] (Figure 1).
The overlap between the physiological responses involved in PTI and ETI partially brings together plant immune signaling pathways in response to biological stresses. The transcriptional regulation of downstream signaling cascades involved in ETI and PTI is controlled by molecules such as small RNAs, transcription factors, chromatin remodelers, histone modifiers (like methylase and acetylase), and transcriptional regulatory complexes [27][28] (Figure 1).
2.1. PAMP (PTI) and Effector (ETI)-Stimulated Immunity in Plants
The plant immune system is a complex network of active and passive systems that withstand pathogen invasion of a host [29]. Most plant surface barriers, such as wax coatings, rigid cell walls, cuticular lipids [30], antimicrobial enzymes [31], or secondary metabolites [7], stop pathogens from getting into the plant and causing disease symptoms to appear.
PAMPs/apoplastic effectors, intracellular effectors, or changed effector targets are actively detected by receptors in the plasma membrane or R proteins in the cytoplasm, activating PTI and ETI. Helper proteins and guard proteins/decoys play a role in pathogen-derived component co-perception [32]. Pathogens use host proteins (S-proteins) expressed by plant sensitivity genes (S-genes) to help enter and multiply, which leads to host adaptation [33]. To get past these protective layers, pathogens use microbial (or pathogen)-associated molecular patterns (MAMP/PAMP)-stimulated immunity (MTI/PTI) and enhanced T-cell immunity (ETI) [30] (Figure 2).
Figure 2. Schematic view of plant intrinsic immunity PAMP-triggered immunity and ETI in plants. The recognition of microbe-dependent molecular patterns (MAMPs) such as flagellin, elongation factor-Tu (EF-Tu), and chitin by cognate sample assessment receptors (PRRs; FLS2, EFR, and CERK) activates many signaling pathways, resulting in the production of reactive oxygen species (ROS), nitric oxide (NO), Ca2+ flux, and the activation of several protein kinases such as CDPKs and MAP kinases (MAPKs). Pathogens release effector molecules into the plant to repress these primary signaling events. However, some plant varieties may know effectors with the aid of R proteins (CC-NB-LRR and TIR-NB-LRR) to compel a hypersensitive response (HR) and systemic acquired resistance (SAR). Targets that are phosphorylated include the replication agents WRKY33, WRKY8, WRKY29, VIP1, MYB51, MYB34, and ERF104, and the chromatin remodeling factors HD2B, GCN5, and SMC1/3. This activity has a pattern of no single transcriptional reprogramming and infusion of primary advocacy-relevant genes but limits pathogen infection and priming plants versus subsequence attacks. Endogenous phytohormones, such as salicylic acid (SA), jasmonic acid (JA), and ethylene, are also induced and contribute to plant immunity.
2.2. Systemic Propagation of Immunity in Plants
Plants react to their surroundings in a stimulus-specific manner. These reactions frequently converge on multiple phytohormone pathways interacting with one another and presumably fine-tune plants' pathway responses to optimize fitness [34][35]. PTI and ETI are both SA-dependent and elicit a systemic, SA-dependent defense response known as systemic acquired resistance (SAR) [2]. SAR is a type of long-term resistance to a wide range of (hemi-) biotrophic infections [36]. Azelaic acid and glycerol-3-phosphate are also considered as additional signals in SAR [37]. Another common type of induced (pathogen) resistance is induced systemic resistance (ISR), which is triggered by commensal bacteria in the plant's rhizosphere [38]. ISR appears to work against a broader spectrum of pathogens than SAR, including both (hemi-) biotrophic and necrotrophic infections. Systemic immunity similar to SAR has been seen in monocots such as maize, barley, wheat, and banana [39][40]. Wheat systemic resistance against Xanthomonas translucens is not related to SA. However, it may be linked to JA/ABAPip (pipecolic acid) and/or WRKY and ETHYLENE RESPONSE FACTORS, which build up in barley petiole exudates after an infection that makes the plant resistant [40] (Figure 2). Systemic immunity against Blumeria graminis f.sp. hordei may depend on SA, whereas Pip induces systemic resistance in barley against both infections [41]. Pip treatment induces NO buildup and primes ROS generation [41], indicating that the Pip pathway of SAR may be conserved between monocotyledonous and dicotyledonous plants (Figure 3). Understanding the similarities and differences between induced resistance responses in monocots and dicots is important. Before using SAR signaling components like those from Arabidopsis to protect cereal crops, it will be important to study SAR in monocots such as barley and wheat.
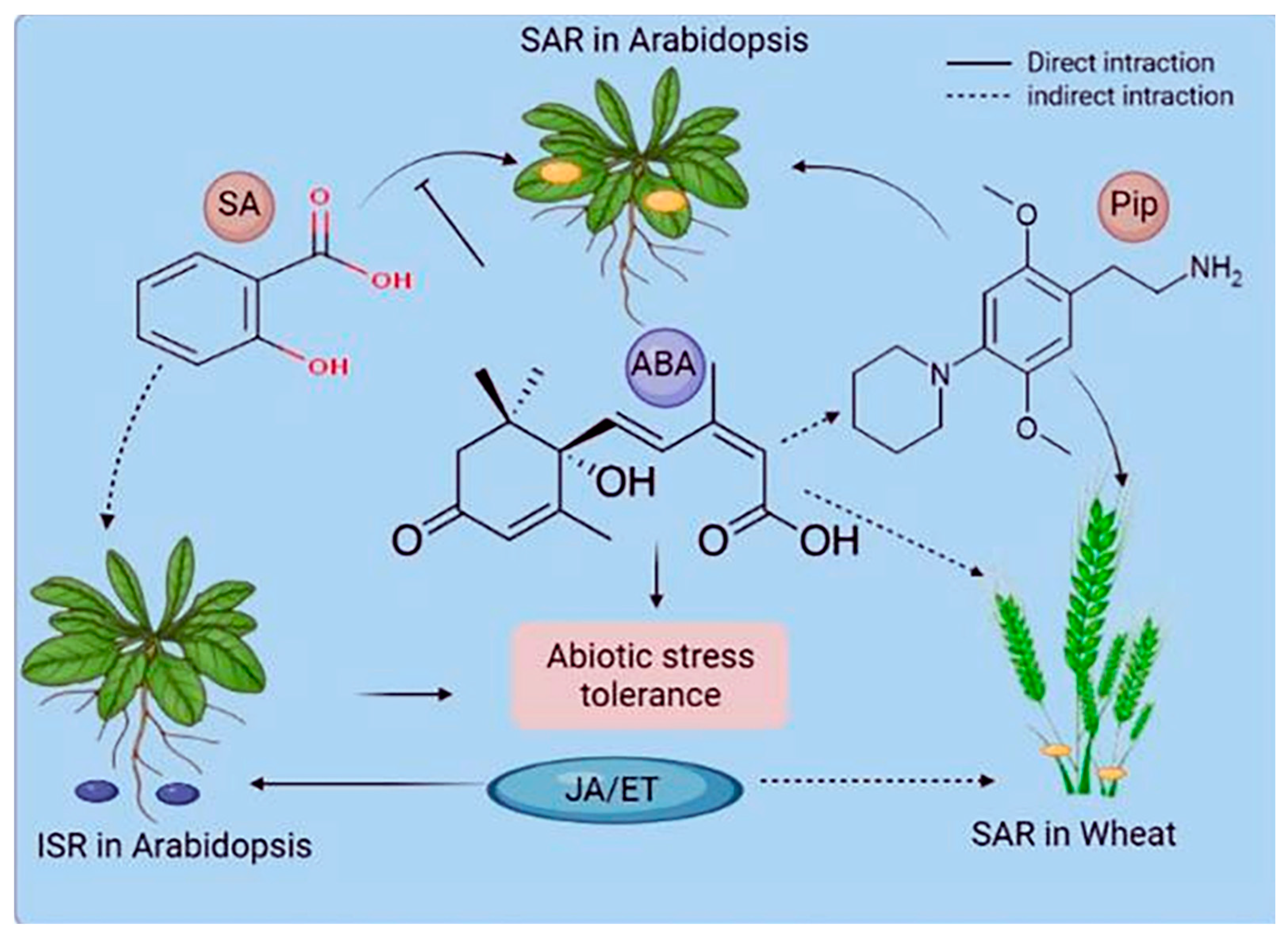
Figure 3. Phytohormones' role in systemic acquired resistance (SAR) and induced systemic resistance (ISR). The interactions of stress-associated phytohormones vary based on the systemic resistance mechanism and the plant species. Hormones can directly function in signaling or pathway antagonism or have an indirect or undefined role. ABA denotes abscisic acid, JA denotes jasmonic acid, ET denotes ethylene, and Pip denotes pipecolic acid.
3. Genetic Effects on Plant Immunity
3.1. Signaling Pathways Host Innate Immunity in Plants
Phytopathogens such as biotrophs, hemibiotrophs, and necrotrophs stress the development of their host plants, reducing yields. Plants are constantly in contact with such phytopathogens, making them vulnerable to their attack. Plants must evolve immunity to these attacks to defend themselves. As a result, plants have evolved ways of recognizing and combating disease via PTI and ETI [42].
3.1.1. Pattern-Triggered Immunity
The first step in the immune response is PTI. PAMPs or MAMPs are identified by PRRs and conserved molecular patterns such as lipopolysaccharides, peptidoglycans, chitin, flagellin, EF-Tu, DNA, and ergosterol. The production of ROS, the activation of MAP kinase, and the stimulation of pathogen-responsive gene transcription are some of the well-known MAMP evaluations included in the activation of signaling pathways and hydrolysis [43][44].
3.1.2. Effector-Triggered Immunity
Plants, including R proteins, are able to recognize many effectors released by pathogens to activate their defense systems. ETI is detected by nucleotide-binding oligomerization-like receptors (NOD), which target microorganism-produced effector molecules [45]. The complex network of cell walls and their key protein constituents is one of the most critical aspects of the plant's immune system. Significant advances in research on how plant viruses affect cell wall remodeling in both susceptible and resistant plants show that cell wall metabolism components can change virus transmission and turn on apoplast- and symplast-based defense mechanisms [46].
3.2. Signal Transduction
3.2.1. ABA Signaling Pathway
ABA is a plant defense-related phytohormone that positively or negatively regulates the immune system, playing a central role in response to abiotic and biotic stresses. The ABA signaling pathway modulates plant growth and development, seed dormancy and maturation, vegetative growth, and stomatal closure in response to various abiotic stimuli. The ABA signaling pathways have been described from hormone receptors to multiple downstream components as a major system that mediates different abiotic stress responses to drought, osmotic, and salt challenges [47][48]. ABA can be synthesized from farnesyl diphosphate (direct C15 pathway) or 9-cis-violaxanthin (indirect C40 pathway). Plants have developed various defense mechanisms to adapt to environmental changes [49]. Under stress, plants accumulate ABA, which serves as a communication molecule among plant cells and initiates various defense mechanisms [50]. Most ABA is synthesized as 9-cis-violaxanthin. In plastids, carotenoids are catabolized into zeaxanthin, which is converted to lutein by zeaxanthin epoxidase. Lutein is converted to all-trans-violaxanthin, which is transformed into 9-cis-violaxanthin or all-trans-neoxanthin; 9-cis-violaxanthin is derived from all-trans-neoxanthin [51]. Under aerobic conditions, 9-cis-epoxycarotenoid dioxygenase converts 9-cis-violaxanthin into 9-cis-neoxanthin and xanthin, which are transferred from the plastid into the cytoplasm [52]. Oxyxanthin is converted into xanthic acid, which serves as a substrate for short-chain alcohol dehydrogenase to generate ABA and abscisic acid. Abscisic aldehyde is further converted into ABA by aldehyde dehydrogenase [53]. Thus, ABA's binding to its receptor initiates various signaling cascades to modulate different cellular processes, including gene expression. In plants, abiotic stimuli induce activation of MAPKs and SNF1-related protein kinase 2 (SnRK2) family members, which play a central role in osmotic stress responses. In Arabidopsis, osmotic stress and ABA activate all SnRK2 family members except SnRK2.9 [54]. The activities of SnRK2 family members, osmotic stress-activated protein kinase, and SAPK1 in Nicotiana tabacum were inhibited by phosphatase treatment, suggesting the importance of phosphorylation in activating SnRK2 proteins [55]. Numerous phosphorylation sites in SnRK2 have been identified. The serine at position 175 in SnRK2.6 has been extensively characterized and proven essential for SnRK2.6 activation [56][57]. Furthermore, ABA treatment increased in vivo phosphorylation levels [58]. NO-induced SnRK2.6 modification inhibits the activity of SnRK2.6. It has been suggested that NO induction by ABA inactivates SnRK2.6 in guard cells, serving as a negative feedback loop that regulates SnRK2.6 activity in response to ABA pathway activation [59]. Phosphatidic acid is a signaling lipid involved in various stress-induced pathways. McLoughlin et al. [60] found that SnRK2.4 and SnRK2.10 accumulated on the cell membrane by binding to phosphatidic acid, promoting salt resistance and root growth and morphogenesis under salt stress. SnRK2s have been shown to bind to ABA-responsive element-binding factors (ABFs), which, upon phosphorylation, regulate the expression of numerous ABA-induced genes [61]. In wheat, PKABA1 is the phosphorylation substrate of TaABF, a seed-specific ABF [55]. In rice, the serine at position 102 of the ABF TRAB1 is crucial for SAPK8/9/10 activation, regulating downstream targets of the ABA signal transduction pathway in response to osmotic stress [62]. In Arabidopsis, ABF1, AREB1/ABF2, AREB2/ABF4, and ABF3 are the most important transcription factors downstream of SnRK2.2/2.3/2.6 in the ABA pathway during the osmotic stress response [61]. These findings highlight the crucial role of SnRK2s in regulating ABA response gene expression by phosphorylating ABFs. SnRK2s also activate the slow anion channel-associated 1 (SLAC1) protein, which regulates ion balance and controls stomatal closure in response to ABA accumulation [63].
3.2.2. Gibberellic Acid (GA) Signaling Pathway
GA is a tetracyclic diterpenoid belonging to the gibberellin family. It is involved in plant growth, metabolism, and response to abiotic stress [52]. The biosynthetic pathways of GA have been extensively investigated [64]. Ent-copalyl diphosphate synthase, ent-kaurene synthase, ent-kaurene oxidase, ent-kaurenoic acid oxidase, GA13-oxidase, GA20-oxidase (GA20ox), GA3-oxidase (GA3ox), and GA2-oxidase are enzymes essential for GA biosynthesis [65]. Geranylgeranyl diphosphate is converted into the endogen ent-kaurene by ent-copalyl diphosphate synthase and ent-kaurene synthase in plastids. In the endoplasmic reticulum, GA12-aldehyde is formed by ent-kaurene oxidation.
In the cytoplasmic matrix, GA12 and GA53 are oxidized by GA20ox and GA3ox to form different GAs [66] (Figure 4). GA-insensitive and topless-related members of the DELLA (aspartic acid–glutamic acid–leucine–leucine–alanine) protein family and negative regulators of GA signal transduction bind to the promoter of GA20ox after recruitment by the transcription factor GAF1, inhibiting GA biosynthesis. Wu et al. [67] reported the interaction between the zinc finger protein OsLOL1 and the transcription factor OsbZIP58, promoting GA biosynthesis. Serine/threonine phosphorylation has been shown to play an essential role in GA synthesis, as dephosphorylation of the repressor of GA1 induced the transcription of GA20ox and GA3ox [68] (Figure 4).
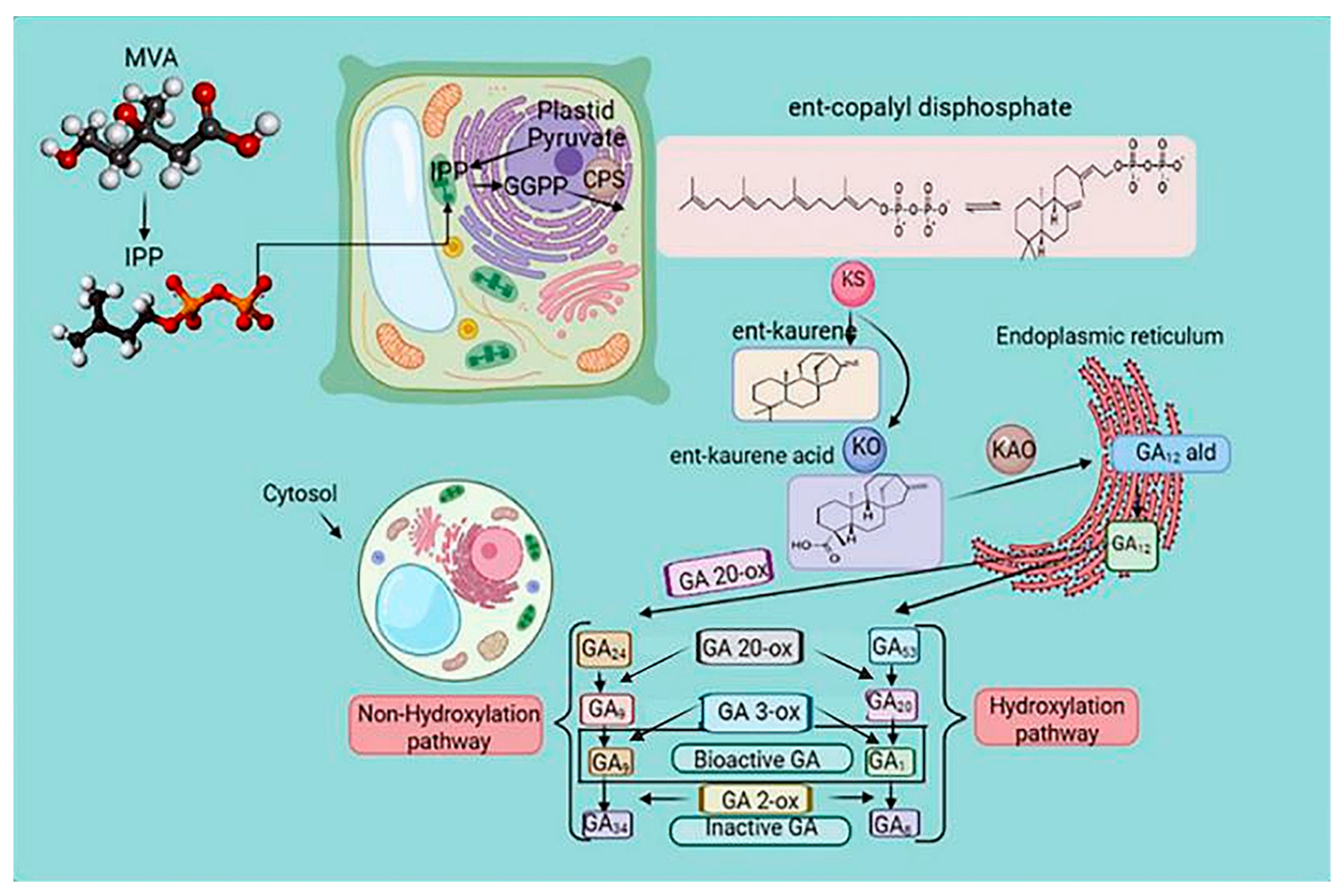
Figure 4. The gibberellin biosynthesis pathway in higher plants. The cellular location of metabolites in plastids is the endoplasmic reticulum and cytoplasm. Methylerythritol phosphate (MEP) and mevalonic acid (MVA) pathways regulate the biosynthesis of gibberellic acid (GA). Ent-copalyl diphosphate synthase (CPS) catalyzed the conversion of geranylgeranyl diphosphate (GGPP) to the endogen ent-kaurene and ent-kaurene synthase (KS), which is the precursor of GA. The biosynthesis of GA is a process of various enzymatic reactions, and the oxidation reaction of GAox is the critical enzymatic reaction. Arrows indicate a cascade reaction catalyzed by an enzyme. Gibberellin Biosynthetic Pathway in Higher Plants. GGPP, trans-genanylgeranyldiphosphate; CDP, ent-copalyl diphosphate; CPS, CDP synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase; GA20ox, GA 20-oxidase; and GA3ox, GA 3-oxidase.
Plants sense extracellular GA via the soluble GA receptor GID1. The binding of GA to GID1 is concentration-dependent. At low extracellular GA concentrations, GA does not bind to GID1, and the N-terminal extension domain of GID1 binds to GA-responsive genes, suppressing their transcription. When the concentration of extracellular GA is high, GID1 binds to GA via its C-terminal domain, inducing the transcription of GA-responsive genes. GA binding to GID1 leads to a conformational change in GID1, and the N-terminal extension domain of GID1 forms a hydrophobic surface that binds to DELLA proteins. GA–GID1 complexes bind to DELLA proteins, forming trimers, which bind to the ubiquitin E3 linker complex (SCFSLY1/GID2), leading to DELLA protein degradation by the 26S proteasome [69]. In the absence of GA, GID1 binds to DELLA proteins, activating alternative pathways [70]. Therefore, plants can sense extracellular GA via both GA-dependent and GA-independent pathways. Ca2+, cyclic guanosine monophosphate (cGMP), and NO are critical second messengers in GA signal transduction [71]. These second messengers transform extracellular cues into intracellular signals, triggering various physiological responses. Ca2+ and cGMP are positive regulators of GA signal transduction, whereas NO is a negative regulator of the GA signal pathway. In rice, Ca2+ binds to calmodulin to form Ca2+/calmodulin, which regulates the expression of Ca2+/ATPase-dependent GA-responsive genes [72]. In Arabidopsis, cGMP and the GA response element play essential roles in the GA signaling pathway, modulating the expression of negative regulators of the GA pathway, SPINDLY, and GID1. Lozano-Juste and Leon [71] confirmed that NO reduced the expression levels of SLY (SLEEPY) and promoted the accumulation of DELLA proteins, thereby inhibiting the expression.
3.2.3. ET Signaling Pathway
ET biosynthesis is induced in PTI and ETI, and ET induction is enhanced by SA [39]. ET is involved in response to various abiotic stresses in plants. For instance, low-temperature stress changes the endogenous level of ET. In Arabidopsis seedlings, ET treatment decreased resistance to frost stress, whereas treatment with ET synthesis inhibitors increased resistance to frost stress. This suggests that ET negatively regulates tolerance to low temperatures [73].
In contrast, ET increased the frost resistance in Arabidopsis seedlings grown in soil [74]. The effect of ET on frost resistance may vary depending on the growth conditions [75]. Under salt stress, ET negatively regulated salt tolerance in Arabidopsis. Overexpression of the wheat TaACO1 (aminocyclopropane-1-carboxylate oxidase) gene reduced two crucial transcription factors, DREB1B/CBF1 and DREB1A/CBF3, suggesting the decrease in stress tolerance in Arabidopsis was due to increased salt sensitivity [76]. Although ET promotes stomatal opening by inhibiting ABA-induced stomatal closure, it also promotes stomatal closure by inducing ROS production in guard cells [77]. Methionine-derived 1-aminocyclopropane-carboxylic acid is the primary ET precursor. During the Yang cycle, methionine is converted to S-adenosylmethionine by methionine adenosyltransferase. The ET precursor 1-aminocyclopropane-1-carboxylic acid is formed by 1-aminocyclopropane-1-carboxylic acid synthetase, and 1-aminocyclopropane-carboxylic acid is converted to ET by 1-aminocyclopropane-1-carboxylic acid oxidase. Methionine is replenished by 5-methylthioadenosine, produced simultaneously as 1-aminocyclopropane-carboxylic acid [78] (Figure 5). The ET signaling pathway has been extensively evaluated in Arabidopsis. The ET receptor (ETR) is located on the surface of the endoplasmic reticulum. In the absence of ET, ETR inhibits activation of the ET signaling pathway by binding to CTR. However, ET binds to ETR, releasing the CTR protein in the presence of ET in the cell. Subsequently, CTR activates EIN2 by cleavage; the carboxyl end of EIN2 enters the nucleus, activating the transcription of EIN3, and the resulting EIN3 protein induces ERF expression [79] (Figure 5).
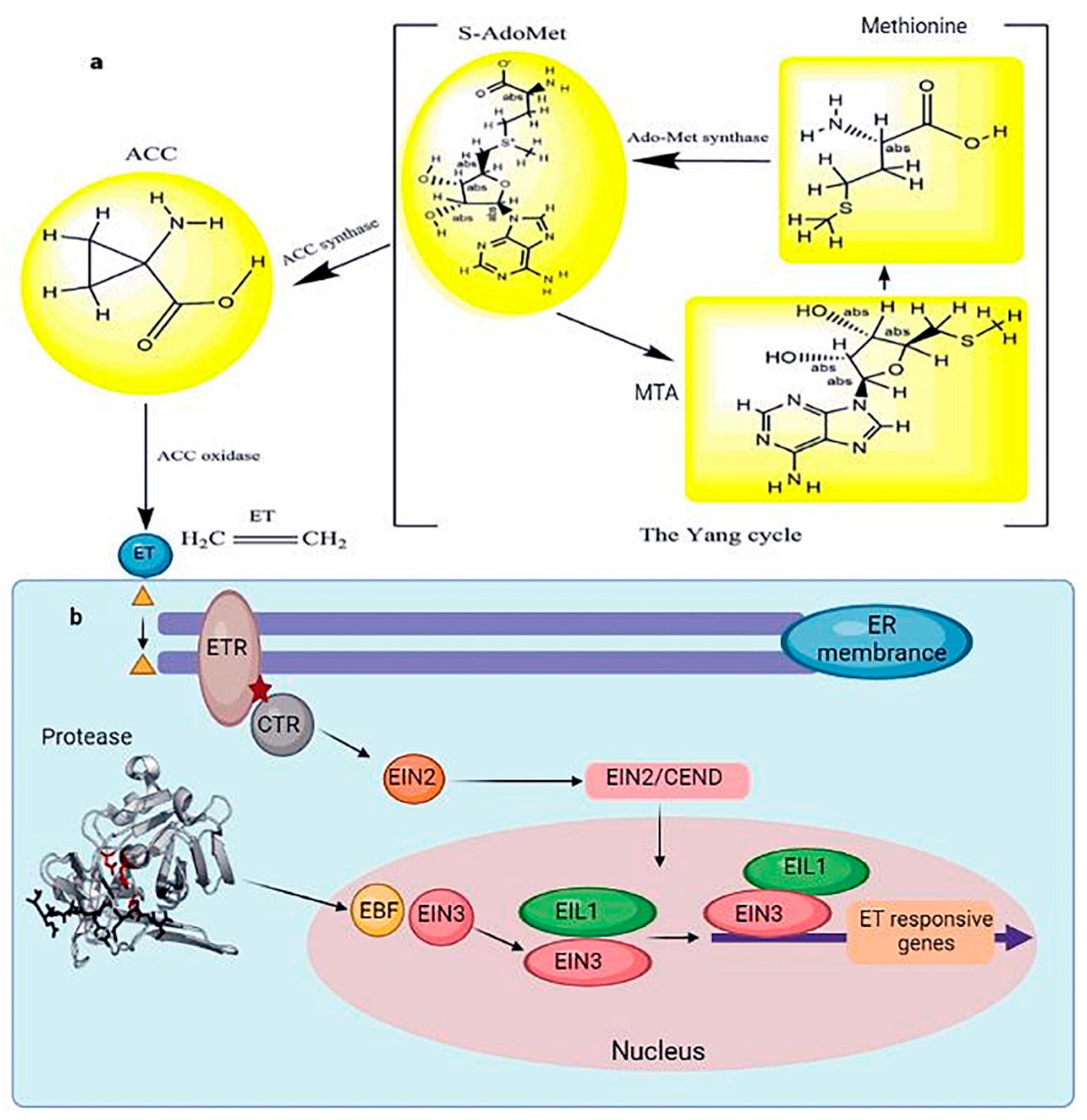
Figure 5. (a) The schematic Yang cycle exhibits the biosynthesis of ET in higher plants. In many cells, methionine produces S-adenosylmethionine under the catalysis of the methyl donor methionine adenosyltransferase. Under the catalysis of 1-aminocyclopropane-1-carboxylic acid synthase, the ET precursor 1-aminocyclopropane-1-carboxylic acid is generated. 1-aminocyclopropane-1-carboxylic acid oxidase catalyzes the production of ET from ACC. (b) ET signal transduction in response to abiotic stress. At the same time, 5 methylthioadenosine, also known as MTA, is produced. Methylthioadenosine, in turn, can be utilized to produce new methionine. An enzyme cascade reaction is denoted by an arrow when an enzyme catalyzes it. When ET binds to ETR, it causes the CTR protein to be released. The main controllers of ethylene signaling that support a range of plant responses to ethylene are the transcription factors EIN3/EIL1. The signaling outcome of ethylene, which controls various characteristics of plant development and stress responses, must be modulated correctly according to the spatiotemporal and environmental circumstances. CTR activates EIN2 by degradation, and the carboxyl end of EIN2 translocates into the nucleus and activates EIN3 transcription. Subsequently, EIN3 induces ERF expression, inhibiting ET-responsive gene expression. Bars represent inhibition, and arrows represent activation.
EIN3 is a nuclear protein acting as a plant-specific transcription factor. It plays a critical role in the ET signaling pathway by directly regulating the expression of ET-responsive genes. Arabidopsis has five EIN3 homologous genes (EIL1–5), and EIN3/EIL1 controls the expression of most ET-responsive genes, as reflected by the finding that EIN3/EIL1 defects result in ET insensitivity. Although ET treatment does not alter EIN3 mRNA levels, it causes the accumulation of the EIN3 protein [80]. In the absence of ET, EIN3 binds to the F-box protein EBF, which results in its recruitment to the Cullin protein complex for ubiquitination. Therefore, the level of EIN3 protein in plants is deficient under physiological growth conditions. However, in the presence of ET, a portion of EIN2 translocates into the nucleus, promoting EBF degradation and subsequent EIN3 stabilization [7][81]. Numerous studies have shown that the EIN3/EIL complex promotes salt stress resistance [65] and low-temperature stress [73]. EIN3/EIL1 directly regulates the expression of the ERF transcription factor in plants, and ERF regulates the amount of ET-responsive genes produced in response to abiotic stresses [82][83] (Figure 5).
3.2.4. SA and JA Signaling Pathways
Important roles in plant defense are played by the phytohormones SA and JA. Various studies have shown that SA- and JA-mediated signals interact among themselves (SA-JA crosstalk) to regulate plant innate immunity against pathogens. SA and JA, which are antagonistic defensive hormones involved in defense against biotrophs and necrotrophs, accumulate at high levels during ETI [84]. The plant hormones SA and JA function as critical secondary signaling molecules in plant immunity [39]. In response to plant-pathogen interactions, primary signaling pathways influence the levels and activities of SA and JA [34][85]. Notably, the treatment of cucumber seedlings with SA synthesis inhibitors blocked the accumulation of endogenous SA and rendered plants more susceptible to the damage caused by low temperatures. Inhibition of SA synthesis under low-temperature stress led to the accumulation of hydrogen peroxide, highlighting the importance of SA in response to low-temperature stress by regulating the expression of low-temperature-responsive genes and intracellular hydrogen peroxide levels [86]. JA is involved in tolerance to low temperatures and freezing [75]. Notably, exogenous JA increased frost resistance in Arabidopsis, while mutations in JA biosynthesis genes increased sensitivity to freezing [87]. In addition, JA has also been implicated in salt tolerance. Specifically, the branch of the JA synthesis pathway catalyzed by allene oxide cyclase via MYC2-dependent and ABA-independent pathways is responsible for the JA-mediated increased salt tolerance in plants [88].
3.2.5. Crosstalk in Stress Response Signaling Pathways
Abiotic stresses, such as drought, salinity, and low and high temperatures, initiate systematic responses in plants. Biological crosstalk refers to instances in which one or more components of one signal transduction pathway affect another. This crosstalk can be achieved through various mechanisms, including the interactions among different signaling cascade protein components. In plants, however, stress response pathways are not initiated independently, and apart from local responses, local stress also triggers systemic responses. For example, local salt stress at the root tip can alter calcium and ROS levels in the whole plant, affecting salt tolerance systemically [89]. The zinc finger protein ZAT12 is involved in DNA repair and replication, essential in ensuring cell survival by acclimation to different environmental stress conditions via RBOHD-mediated ROS accumulation [90].
Activating the intracellular Ca2+ signal enhances ROS production and the expression of defense genes, initiating systemic plant defense responses [91]. The guard cell hydrogen-peroxide-resistant 1 (GHR1) protein is a plasma membrane receptor kinase expressed in guard cells and regulates the activation of plasma membrane calcium channels [92]. It has been reported that JA responses positively relate to SA [93] and ET [94] responses because of synergistic or feedback regulation. In Arabidopsis, EIN3/EIL1 is regulated by EBF1/2. EIN3/EIL1 regulates the transcriptional repressor jasmonate ZIM domain (JAZ) and causes it to activate ET-responsive gene expression.
Additionally, JAZ inhibits the transcriptional activity of EIN3/EIL1, suppressing the expression of the Erf1 gene. MeJA, a vital component of the JA pathway, increased ET levels in plants [95]. Although JA and ET both positively regulate plant defense responses, they may also have antagonistic effects [96]. Studies have shown that EIN3/EIL1 levels are also regulated by transcription factors activated by other hormones, such as GA. DELLA proteins, transcriptional repressors of the GA signaling pathway, bind to regulatory sequences in Ein3/Eil1 genes, suppressing their transcription [97]. However, in the absence of GA, DELLA and MYC2 bind to JAZs, and the binding of MYC2 to G-box motifs in JA-responsive genes activates their expression. When the GA content increases, DELLA proteins are degraded, and JAZs are released, inhibiting the JA signaling pathway [98]. Yaish et al. [99] demonstrated that OsAP2-39 overexpression induced the upregulation of 9-cis-epoxycarotenoid dioxygenase I, a key enzyme in ABA biosynthesis. OsAP2-39 also plays an essential role in maintaining the balance between ABA and GA in plants by regulating the expression of critical enzymes involved in ABA and GA biosynthesis. In response to abiotic stress in plants, DELLA proteins participate in the dynamic balance between ABA and GA by enhancing the expression of the ubiquitin E3 ligase Xerico, reducing GA levels, and increasing ABA levels [100].
References
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320.
- Spoel, S.H.; Dong, X.N. How do plants achieve immunity?: Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100.
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621.
- Holmes, E.C.; Chen, Y.C.; Sattely, E.S.; Mudgett, M.B. An engineered pathway for N-hydroxy-pipecolic acid synthesis enhances systemic acquired resistance in tomato. Sci. Signal. 2019, 12, eaay3066.
- Lee, S.; Whitaker, V.M.; Hutton, S.F. Mini Review: Potential Applications of Non-host Resistance for Crop Improvement. Front. Plant Sci. 2016, 7, 997.
- Salguero-Linares, J.; Coll, N.S. Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 2019, 70, 2087–2095.
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674.
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Genome-Wide Characterization and Abiotic Stresses Expression Analysis of Annexin Family Genes in Poplar. Int. J. Mol. Sci. 2022, 23, 515.
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14.
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58.
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875.
- Zhu, M.; Shabala, L.; Cuin, T.A.; Huang, X.; Zhou, M.; Munns, R.; Shabala, S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 2016, 67, 835–844.
- Tsuda, K.; Sato, M.; Glazebrook, J.; Cohen, J.D.; Katagiri, F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008, 53, 763–775.
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844.
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227.
- Pieterse, C.M.; Leon-Reyes, A.; van der Ent, S.; van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316.
- Navarro, L.; Jay, F.; Nomura, K.; He, S.Y.; Voinnet, O. Suppression of the microRNA pathway by bacterial effector proteins. Science 2008, 321, 964–967.
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; et al. RRM Transcription Factors Interact with NLRs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol. Cell 2019, 74, 996–1009.e7.
- Albert, I.; Hua, C.; Nürnberger, T.; Pruitt, R.N.; Zhang, L. Surface Sensor Systems in Plant Immunity. Plant Physiol. 2020, 182, 1582–1596.
- Wan, W.-L.; Fröhlich, K.; Pruitt, R.N.; Nürnberger, T.; Zhang, L. Plant cell surface immune receptor complex signaling. Curr. Opin. Plant Biol. 2019, 50, 18–28.
- Yu, H.; Zhang, H.; Dong, M.; Wu, Z.; Shen, Z.; Xie, Y.; Kong, Z.; Dai, X.; Xu, B. Metabolic reprogramming and AMPKα1 pathway activation by caulerpin in colorectal cancer cells. Int. J. Oncol. 2017, 50, 161–172.
- Movahedi, A.; Zhang, J.; Sun, W.; Kadkhodaei, S.; Mohammadi, K.; Almasizadehyaghuti, A.; Yin, T.; Zhuge, Q. Plant small RNAs: Definition, classification and response against stresses. Biologia 2018, 73, 285–294.
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539.
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol. Plant 2010, 3, 783–793.
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834.
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network Properties of Robust Immunity in Plants. PLoS Genet. 2009, 5, e1000772.
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant Immunity: From Signaling to Epigenetic Control of Defense. Trends Plant Sci. 2018, 23, 833–844.
- Ding, B.; Wang, G.L. Chromatin versus pathogens: The function of epigenetics in plant immunity. Front. Plant Sci. 2015, 6, 675.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449.
- Habib, H.; Majid, K. Plant protease inhibitors: A defense strategy in plants. BMBR 2007, 2, 68–85.
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959.
- Doehlemann, G.; Hemetsberger, C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 2013, 198, 1001–1016.
- Pieterse, C.M.J.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521.
- Erb, M. Plant Biology: Evolution of Volatile-Mediated Plant-Plant Interactions. Curr. Biol. 2019, 29, R873–R875.
- Luna, E.; Bruce, T.J.A.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853.
- Ádám, A.L.; Nagy, Z.Á.; Kátay, G.; Mergenthaler, E.; Viczián, O. Signals of Systemic Immunity in Plants: Progress and Open Questions. Int. J. Mol. Sci. 2018, 19, 1146.
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375.
- Yang, D.-L.; Yang, Y.; He, Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant 2013, 6, 675–685.
- Dey, S.; Wenig, M.; Langen, G.; Sharma, S.; Kugler, K.G.; Knappe, C.; Hause, B.; Bichlmeier, M.; Babaeizad, V.; Imani, J.; et al. Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 2014, 166, 2133–2151.
- Lenk, M.; Wenig, M.; Bauer, K.; Hug, F.; Knappe, C.; Lange, B.; Timsy; Häußler, F.; Mengel, F.; Dey, S.; et al. Pipecolic Acid Is Induced in Barley upon Infection and Triggers Immune Responses Associated with Elevated Nitric Oxide Accumulation. Mol. Plant Microbe Interact. 2019, 32, 1303–1313.
- Kumar, J.; Ramlal, A.; Kumar, K.; Rani, A.; Mishra, V. Signaling Pathways and Downstream Effectors of Host Innate Immunity in Plants. Int. J. Mol. Sci. 2021, 22, 9022.
- Choi, H.W.; Manohar, M.; Manosalva, P.; Tian, M.; Moreau, M.; Klessig, D.F. Activation of Plant Innate Immunity by Extracellular High Mobility Group Box 3 and Its Inhibition by Salicylic Acid. PLoS Pathog. 2016, 12, e1005518.
- Zhong, M.; Yan, H.; Li, Y. Flagellin: A unique microbe-associated molecular pattern and a multi-faceted immunomodulator. Cell Mol. Immunol. 2017, 14, 862–864.
- Latrasse, D.; Jégu, T.; Li, H.; de Zelicourt, A.; Raynaud, C.; Legras, S.; Gust, A.; Samajova, O.; Veluchamy, A.; Rayapuram, N.; et al. MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol. 2017, 18, 131.
- Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Plant Cell Wall as a Key Player during Resistant and Susceptible Plant-Virus Interactions. Front. Microbiol. 2021, 12, 656809.
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679.
- Joshi-Saha, A.; Valon, C.; Leung, J. A brand new START: Abscisic acid perception and transduction in the guard cell. Sci. Signal. 2011, 4, re4.
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099–1111.
- Zhang, J.H.; Jia, W.S.; Yang, J.C.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119.
- Barrero, J.M.; Piqueras, P.; Gonzalez-Guzman, M.; Serrano, R.; Rodriguez, P.L.; Ponce, M.R.; Micol, J.L. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 2005, 56, 2071–2083.
- Qin, X.; Zeevaart, J.A. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361.
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846.
- Boudsocq, M.; Barbier-Brygoo, H.; Lauriere, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766.
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177.
- Ng, L.M.; Soon, F.F.; Zhou, X.E.; West, G.M.; Kovach, A.; Suino-Powell, K.M.; Chalmers, M.J.; Li, J.; Yong, E.L.; Zhu, J.K.; et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. USA 2011, 108, 21259–21264.
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839.
- Kline, K.G.; Barrett-Wilt, G.A.; Sussman, M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 15986–15991.
- Wang, P.C.; Dua, Y.Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.J.; Zhu, X.H.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618.
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Lauriere, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449.
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27.
- Johnson, R.R.; Wagner, R.L.; Verhey, S.D.; Walker-Simmons, M.K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002, 130, 837–846.
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486.
- Liu, Y.; Xie, M.-X.; Jiang, M.; Wang, Y.-D. Spectroscopic investigation of the interaction between human serum albumin and three organic acids. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2005, 61, 2245–2251.
- Zhang, L.X.; Li, Z.F.; Quan, R.D.; Li, G.J.; Wang, R.G.; Huang, R.F. An AP2 Domain-Containing Gene, ESE1, Targeted by the Ethylene Signaling Component EIN3 Is Important for the Salt Response in Arabidopsis. Plant Physiol. 2011, 157, 854–865.
- Tudzynski, B.; Mihlan, M.; Rojas, M.C.; Linnemannstons, P.; Gaskin, P.; Hedden, P. Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: Des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J. Biol. Chem. 2003, 278, 28635–28643.
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733.
- Wang, W.; Zhang, J.; Qin, Q.Q.; Yue, J.; Huang, B.Y.; Xu, X.F.; Yan, L.F.; Hou, S.W. The six conserved serine/threonine sites of REPRESSOR OF ga1-3 protein are important for its functionality and stability in gibberellin signaling in Arabidopsis. Planta 2014, 240, 763–779.
- Sun, T.P. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010, 154, 567–570.
- Yamamoto, Y.; Hirai, T.; Yamamoto, E.; Kawamura, M.; Sato, T.; Kitano, H.; Matsuoka, M.; Ueguchi-Tanaka, M. A Rice gid1 Suppressor Mutant Reveals That Gibberellin Is Not Always Required for Interaction between Its Receptor, GID1, and DELLA Proteins. Plant Cell 2010, 22, 3589–3602.
- Lozano-Juste, J.; Leon, J. Nitric Oxide Regulates DELLA Content and PIF Expression to Promote Photomorphogenesis in Arabidopsis. Plant Physiol. 2011, 156, 1410–1423.
- Chen, X.; Chang, M.; Wang, B.; Wu, B. Cloning of a Ca(2+)-ATPase gene and the role of cytosolic Ca2+ in the gibberellin-dependent signaling pathway in aleurone cells. Plant J. 1997, 11, 363–371.
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595.
- Catalá, R.; López-Cobollo, R.; Mar Castellano, M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, J. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 2014, 26, 3326–3342.
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229.
- Chen, D.; Ma, X.; Li, C.; Zhang, W.; Xia, G.; Wang, M. A wheat aminocyclopropane-1-carboxylate oxidase gene, TaACO1, negatively regulates salinity stress in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1815–1827.
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 2006, 47, 907–916.
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218.
- Ji, Y.S.; Guo, H.W. From Endoplasmic Reticulum (ER) to Nucleus: EIN2 Bridges the Gap in Ethylene Signaling. Mol. Plant 2013, 6, 11–14.
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677.
- Binder, B.M.; Walker, J.M.; Gagne, J.M.; Emborg, T.J.; Hemmann, G.; Bleecker, A.B.; Vierstra, R.D. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 2007, 19, 509–523.
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007.
- Onate-Sanchez, L.; Anderson, J.P.; Young, J.; Singh, K.B. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007, 143, 400–409.
- Liu, L.; Sonbol, F.-M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099.
- Kazan, K.; Lyons, R. Intervention of Phytohormone Pathways by Pathogen Effectors. Plant Cell 2014, 26, 2285–2309.
- Dong, C.J.; Li, L.; Shang, Q.M.; Liu, X.Y.; Zhang, Z.G. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 2014, 240, 687–700.
- Hu, Y.R.; Jiang, L.Q.; Wang, F.; Yu, D.Q. Jasmonate Regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924.
- Zhao, Y.; Xing, L.; Wang, X.G.; Hou, Y.J.; Gao, J.H.; Wang, P.C.; Duan, C.G.; Zhu, X.H.; Zhu, J.K. The ABA Receptor PYL8 Promotes Lateral Root Growth by Enhancing MYB77-Dependent Transcription of Auxin-Responsive Genes. Sci. Signal. 2014, 7, ra53.
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941.
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009, 2, ra45.
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749.
- Hua, D.; Wang, C.; He, J.; Liao, H.; Duan, Y.; Zhu, Z.; Guo, Y.; Chen, Z.; Gong, Z. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 2012, 24, 2546–2561.
- Shah, J. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 2003, 6, 365–371.
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49.
- Yu, M.M.; Shen, L.; Fan, B.; Zhao, D.Y.; Zheng, Y.; Sheng, J.P. The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol. Technol. 2009, 54, 153–158.
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479.
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927.
- Bastian, R.; Dawe, A.; Meier, S.; Ludidi, N.; Bajic, V.B.; Gehring, C. Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana. Plant Signal. Behav. 2010, 5, 224–232.
- Yaish, M.W.; El-kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice. PLoS Genet. 2010, 6, e1001098.
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
22 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




