Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ruma Dey Ghosh | -- | 1791 | 2022-11-21 05:36:16 | | | |
| 2 | Conner Chen | + 15 word(s) | 1806 | 2022-11-22 07:38:59 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ghosh, R.D. Mechanism of Actions of Long Non-Coding RNAs. Encyclopedia. Available online: https://encyclopedia.pub/entry/35419 (accessed on 07 February 2026).
Ghosh RD. Mechanism of Actions of Long Non-Coding RNAs. Encyclopedia. Available at: https://encyclopedia.pub/entry/35419. Accessed February 07, 2026.
Ghosh, Ruma Dey. "Mechanism of Actions of Long Non-Coding RNAs" Encyclopedia, https://encyclopedia.pub/entry/35419 (accessed February 07, 2026).
Ghosh, R.D. (2022, November 21). Mechanism of Actions of Long Non-Coding RNAs. In Encyclopedia. https://encyclopedia.pub/entry/35419
Ghosh, Ruma Dey. "Mechanism of Actions of Long Non-Coding RNAs." Encyclopedia. Web. 21 November, 2022.
Copy Citation
Long non-coding RNAs (lncRNAs) are master regulators of almost all biological processes. LncRNAs perform their function mostly through different chromatin-based mechanisms such as signals, decoys, guides, and scaffolds in chromatin remodelling. LncRNAs play a major role in genomic imprinting.
long non-coding RNA (lncRNA)
oral squamous cell carcinoma
recurrence
1. Mechanism of Action
In last few decades, revolutionary changes in our understanding of genome regulation have emerged. The fundamental role of long non-coding RNAs (lncRNA) is the synchronisation and regulation of gene/protein expression, thereby resulting in the fine-tuning of each and every physiological processes. Rarely, they contain short open reading frames (ORFs) [1]. The unique feature of lncRNAs is their ability to interact with other RNAs (e.g., mRNA, circRNA, miRNA, and others), DNA, and protein molecules, and consequently, they regulate many different biological processes at different levels in different ways. The formation of secondary RNA structures allows the lncRNAs to play the key role in controlling the adjacent (cis) and very distant (tans) domains in a specific chromatin region (loci) [2][3]. The functions of lncRNAs are very complex, multidimensional, and multifaceted (Figure 1). Still, they remain unclear, and current research fails to explain the sensitivity and specificity that is needed to achieve these lncRNA-mediated interactions, and the regulation of gene expressions are found to be highly cell tissue-specific in normal patients or in any patients with disease conditions like cancer.
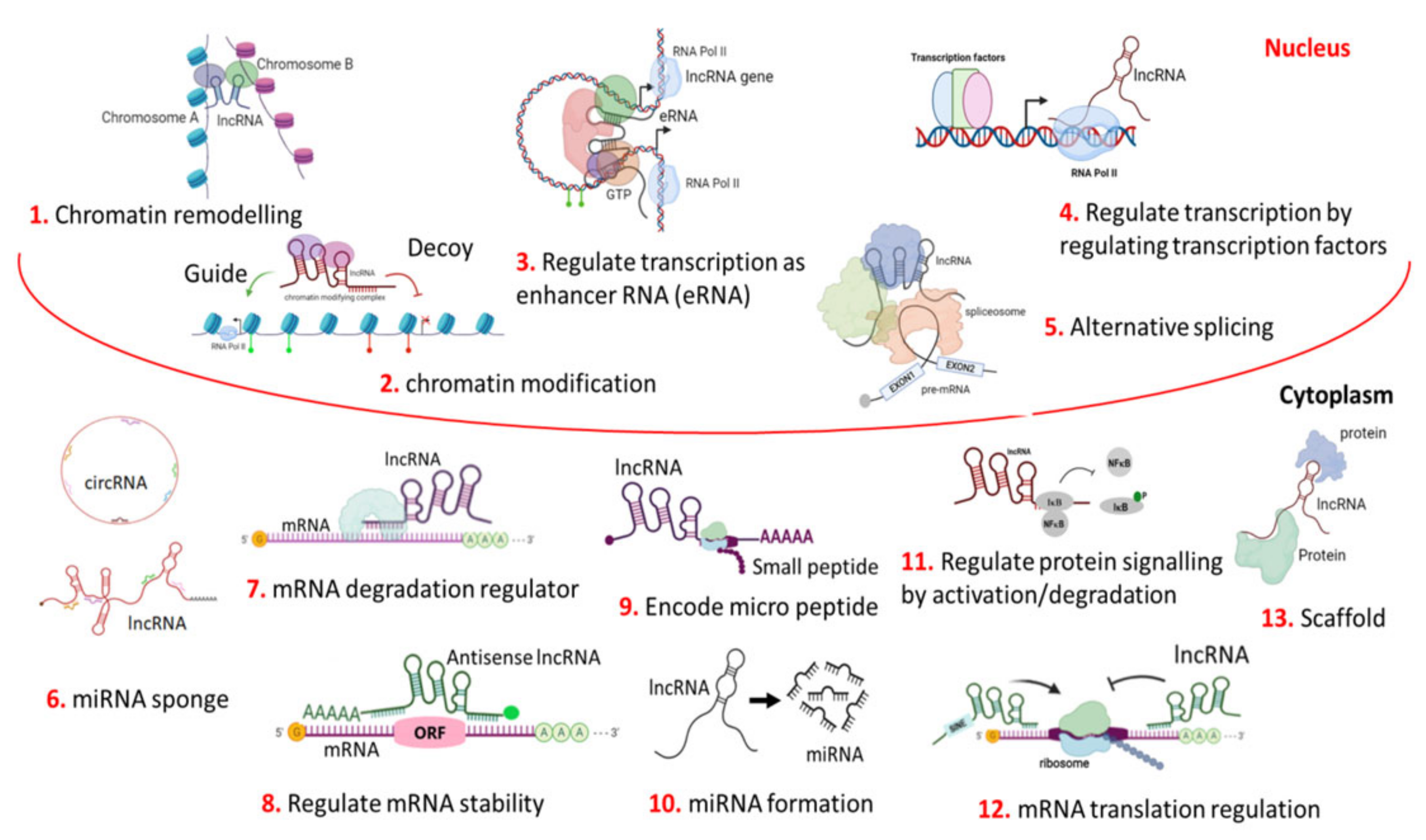
Figure 1. Different, complex, multi-dimensional functions of LncRNA. LncRNA mainly functions as signals, decoys, scaffolds, and as guides. Thus, lncRNA can act as a regulator for chromatin remodelling their environment (neighbouring or distant through intra or inter-chromosomal interaction) by positioning chromatin modifiers at the chromosomal level in the genome architecture (1). LncRNA can regulate gene expression by recruiting a chromatin-modifying complex either to activate or repress the neighbouring genes at the transcription level (2). It can also regulate transcription as an enhancer RNA or eRNA (3), or by the binding and/or activating of transcription factors to the promoter region (4). It can regulate pre-mRNA processing through the alternative splicing of mRNAs (5). In cytoplasm, at a circular form or linear form, it can sponge (silence) the function of miRNA (6). It can also act as an mRNA degradation regulator (7); mRNA stability can also be regulated by lncRNA (8). It can code for small micro peptide (9). They can generate miRNA by degrading themselves (10). It can regulate signalling cascades in different physiological pathways through the activation/deactivation of proteins in cancer cells (11). It can regulate the mRNA translation (12). Architectural scaffolding is another important function of lncRNAs. The LncRNA-mediated scaffolding of protein (RNA-protein/ribonucleoprotein) structures are called paraspeckles, which are found in interchromatin space. This is also found in several shared pathways in the cytoplasm (13).
2. LncRNA as Chromatin Regulators
LncRNAs perform their function mostly through different chromatin-based mechanisms such as signals, decoys, guides, and scaffolds in chromatin remodelling. LncRNAs play a major role in genomic imprinting. For the expression of protein coding genes, lncRNA can regulate their neighbouring (cis) or distant (trans) genomic environments by acting as an enhancer or a diffuser. LncRNA mediates epigenetic modifications by positioning chromatin-remodelling complexes to specific chromatin loci. It was estimated that 38% of lncRNA is found in various tissues binds to PRC2 and to other chromatin-modifying proteins like CoREST (REST corepressor 1 protein encoded by RCOR1 gene) and SMCX (also called JmjC-domain protein encoded by X-linked mental retardation gene SMCX or JARID1C gene) [4]. LncRNAs (ANRIL, XIST (X-inactive specific transcript), KCNQ1OT1, and HOTAIR) bind to trithorax chromatin-activating complexes (TrxG) by recruiting different epigenetic modifiers to its assigned loci for chromatin remodelling [5][6].
ANRIL/CDKN2B, the antisense lncRNA in the INK4 locus, acts as a scaffold and transcriptionally silences the INK4b-ARF-INK4a locus. It binds to polycomb repressive complex 1 and 2 (PRC1 and PRC2) [7][8]. When PRC2 binds to the gene locus, it causes the spreading of the methylation marks, which is distinct for the transcriptional silencing of genes. A 17 Kb gene called X-inactive specific transcript (XIST), that is positioned on the human X chromosome, is responsible for the X chromosome dosage compensation. Cis-X chromosome regulation begins after the X chromosome is covered and PRC2 is recruited to its specific sites, and thus, this results in the emergence of histone H3 lysine K27 trimethylation (H3K27me3) and also causes X-linked inactivation [9][10]. LncRNA HOTAIR (HOX transcript antisense RNA) acts as a scaffold by cleaving itself to PRC2 and mediating the homeobox D cluster (HOXD) locus repression by spreading the H3K27me3 marks, and thus, this causes gene silencing [11]. HOTAIR forms many double stem-loop structures which bind to the lysine-specific demethylase1 (LSD1) and the PRC histone modification complexes [12]. KCNQ1OT1 (KCNQ1-overlapping transcript 1) antisense lncRNA, which belongs to a potassium voltage-gated channel subfamily, remains upregulated in colon cancer [13] and acts as a signal lncRNA by associating itself to G9a histone methyl-transferase and also to PRC2 [14]. When PRC2 and G9a methyl transferase are recruited to KCNQ1OT1, they mediate gene-silencing associated marks such as the demethylation of lysine 9 (H3K9me2) and lysine 27 on histone 3 [15]. KCNQ1OT1 boosts the transcriptional silencing of genes through chromatin remodelling.
3. LncRNA in Transcriptional Regulation
LncRNAs have the potential control in transcriptional regulation through modulating the expression and functions of different transcription factors, which in turn regulates different gene expression. LncRNAs can act as a co-factor of transcription factors and enzymes that are related by chromatin modification. They can regulate gene expression in cis (neighbouring) or in trans (distant) environments. Evf2 lncRNA recruits the transcriptional activator, DLX1, to the key DNA enhancer to repress the gene expression [5][16]. More recently, a detailed study on an ultra-conserved enhancer (UCE) uncovered that it has the lncRNA-dependent topological and transcriptional control, through complex effects, on the chromosome topology by interacting with multi-megabase distant genes. Evf2 lncRNA with Dlx5/6 forms a cloud-forming structure of UCE, which concurrently accomplishes the activation (Umad1, 1.6Mb distant) and repression of (Akr1b8, 27Mb distant) chr6 target genes, locally [5][16]. Recently, a new class of lncRNA, the eRNA (enhancer RNA), has been discovered at the gene enhancer region and is implicated mainly in transcriptional regulation [17].
LncRNA causes interaction with RNA-binding factors, namely heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs then form ribonucleoproteins (RNPs) which then can act as enhancers to promote transcriptional processing by recruiting key transcription machinery proteins to their specific target gene promoters. RNPs can also cause the repression of gene transcription by attaching themselves to existing gene repressors. Fas and Blk are pro-apoptotic genes, for which, lncRNA mediates their repression by acting as a decoy for the transcriptional factor, NFYA [18].
LncRNAs play a major role in the modification of RNA polymerase (RNAP) II activity by interacting with the initiation complex, and guiding it to choose the precise promoter. It has been seen that in humans, the transcription of non-coding RNAs (ncRNAs) from the upstream region of the dihydrofolate reductase (DHFR) locus leads to the formation of a triplex in the promoter region, thus leading to the inhibition of the binding of the transcription factor, TFIID31 [19]. The basic components of RNAP II-dependent transcription machinery interactions with the lncRNA are transcribed by RNAP III. For further regulation, this lncRNA extracts their expression from an RNAP II-dependent transcription reaction. For example, the transcription of Alu elements bind to RNAP II in response to heat shock, eliminating the requirement of the pre-initiation complex and this causes repression by their domain interaction [20].
4. LncRNA in Post-Transcriptional Regulation
LncRNAs are potentially able to recognise their complimentary sequences, thus allowing for interactions that are responsible for the regulation of the post-transcriptional processing of mRNA. The processes in which lncRNAs are involved include capping, splicing, editing, transport, translation, and stability at various control sites. For example, the interaction of MALAT1 with splicing factors interrupts the process of alternate splicing. RNCR2 (also called MIAT or Gomafu) is an lncRNA that affects the mRNA splicing to provide a neuron-specific expression by interacting with the splicing factor 1 (SF1) and blocking the formation of spliceosome [14][20][21]. Natural antisense (NAT) lncRNAs recruit repressor complexes like PRC2 to the target genes and prompt the formation of RNA duplexes, inhibit cis-regulatory elements, and lead to the alternate splicing of paired genes. NAT of ZEB2 binds to the 5′splice site of an intron in 5′ UTR of ZEB2 mRNA. This intron comprises the internal ribosome entry site (IRES), which is the essential component of the translational machinery. NAT overexpression prevents splicing and increases ZEB2 expression and consequently, it down regulates the E-cadherin expression.
5. Role of lncRNA in Genomic Imprinting
Genomic imprinting is a normal epigenetic process of gene regulation by which a subset of genes can be expressed in a parent-of-origin-specific manner from one of the parental chromosomes [20]. Specific genomic loci, known as imprinting control regions (ICRs), control the genomic imprinting. Methylated and unmethylated DNA genomic imprinting regions are dependent on their parental origin for the specific expression of lncRNA genes which leads to the activation or suppression of neighbouring genes in cis-regulating machinery. Instead of PRC2, DNA methyltransferase plays a major role in lncRNA-mediated histone modification and DNA methylation in uniparental gene expression [22]. LncRNA and protein-coding genes are associated with imprinted clusters and are inversely expressed. AIRN and KCNQ1OT1 are examples of such lncRNAs that are responsible for the genomic imprinting of paternally inherited genes [23]. KCNQ1OT1 takes a crucial step in the long-range bidirectional repression of chromatin structures of different protein-coding genes by associating with the chromatin-modifying complexes, EED and G9A/EHMT2, and with the RNA itself. KCNQ1OT1/LIT1 is considered as an imprinted control region 2 (ICR2), which consists of at least eight genes that are expressed from the maternal allele [24]. KCNQ1OT1 silences the KCNQ1 imprinting control region by functioning like an organiser and by interacting with chromatin-modifying complexes, EED and G9A/EHMT2, and with the RNA itself [24][25][26][27][28]. Insulin-like growth factor-2 (Igf2) and insulin-like growth factor-2 receptor (Igf2r) are examples of maternally and paternally imprinted genes [29]. H19, an lncRNA plays an important role in regulating maternal imprinting which is essential for the regulation of cellular differentiation during embryogenesis in humans [30]. H19, after associating with methyl-CpG-binding-domain protein1 (MBD1), recruits histone-lysine-methyl transferase-containing complexes which form repressive H3K9 methylation marks on the targeted imprinting loci. The absence of H19-mediated maternal imprinting may cause Beckwith-Wiedemann Syndrome (BWS) and correlates with an increased risk of developing a Wilms tumour of the kidney [31][32][33][34]. The dysregulation of imprinting genes is reported in some pathological conditions, including cancer [35]. Collectively, the above information has revealed that the functions of lncRNAs are unique, due to their ability to establish molecular interactions with proteins and all types of nucleic acids to modulate/regulate their accessibility, localisations, and functions. The multidimensional and multifaceted functional versatility and flexibility of lncRNAs has emerged in recent days [36][37]. The complete understanding of the functional plasticity of lncRNAs will give fundamental insight into the mechanisms that lncRNAs employ for gene/protein regulation and the pathophysiological processes of cancers, including oral squamous cell carcinoma (OSCC).
References
- Leng, F.; Miu, Y.Y.; Zhang, Y.; Luo, H.; Lu, X.L.; Cheng, H.; Zheng, Z.G. A micro-peptide encoded by HOXB-AS3 promotes the proliferation and viability of oral squamous cell carcinoma cell lines by directly binding with IGF2BP2 to stabilize c-Myc. Oncol. Lett. 2021, 22, 697.
- Sanchez Calle, A.; Kawamura, Y.; Yamamoto, Y.; Takeshita, F.; Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018, 109, 2093–2100.
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055.
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719.
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016, 1, 43–50.
- Kapranov, P.; St Laurent, G.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010, 8, 149.
- Aguilo, F.; Zhou, M.M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369.
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962.
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafreniere, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542.
- Yang, C.; Chapman, A.G.; Kelsey, A.D.; Minks, J.; Cotton, A.M.; Brown, C.J. X-chromosome inactivation: Molecular mechanisms from the human perspective. Hum. Genet. 2011, 130, 175–185.
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076.
- Nakano, S.; Murakami, K.; Meguro, M.; Soejima, H.; Higashimoto, K.; Urano, T.; Kugoh, H.; Mukai, T.; Ikeguchi, M.; Oshimura, M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006, 97, 1147–1154.
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246.
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196.
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756.
- Kim, T.K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622.
- Clemson, C.M.; McNeil, J.A.; Willard, H.F.; Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996, 132, 259–275.
- Sado, T.; Hoki, Y.; Sasaki, H. Tsix silences Xist through modification of chromatin structure. Dev. Cell 2005, 9, 159–165.
- Wan, L.B.; Bartolomei, M.S. Regulation of imprinting in clusters: Noncoding RNAs versus insulators. Adv. Genet. 2008, 61, 207–223.
- He, Y.; Meng, X.M.; Huang, C.; Wu, B.M.; Zhang, L.; Lv, X.W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27.
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499.
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Mattick, J.S.; Gagen, M.J. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 2001, 18, 1611–1630.
- Guo, H.; Wu, L.; Yang, Q.; Ye, M.; Zhu, X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene 2015, 554, 114–119.
- Ishibashi, M.; Kogo, R.; Shibata, K.; Sawada, G.; Takahashi, Y.; Kurashige, J.; Akiyoshi, S.; Sasaki, S.; Iwaya, T.; Sudo, T.; et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol. Rep. 2013, 29, 946–950.
- Li, W.; Zheng, J.; Deng, J.; You, Y.; Wu, H.; Li, N.; Lu, J.; Zhou, Y. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology 2014, 146, 1714–1726.e1715.
- Mattick, J.S.; Amaral, P.P.; Dinger, M.E.; Mercer, T.R.; Mehler, M.F. RNA regulation of epigenetic processes. Bioessays 2009, 31, 51–59.
- Kanduri, C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim. Biophys. Acta 2016, 1859, 102–111.
- Thorvaldsen, J.L.; Duran, K.L.; Bartolomei, M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998, 12, 3693–3702.
- Brunkow, M.E.; Tilghman, S.M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991, 5, 1092–1101.
- DeBaun, M.R.; Tucker, M.A. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J. Pediatr. 1998, 132, 398–400.
- Heery, R.; Finn, S.P.; Cuffe, S.; Gray, S.G. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers 2017, 9, 38.
- Sparago, A.; Cerrato, F.; Vernucci, M.; Ferrero, G.B.; Silengo, M.C.; Riccio, A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 2004, 36, 958–960.
- Jelinic, P.; Shaw, P. Loss of imprinting and cancer. J. Pathol. 2007, 211, 261–268.
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206.
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407.
More
Information
Subjects:
Health Care Sciences & Services
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
22 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




