In cyanobacteria and some algae, FMN-containing flavodoxin (Fld) acts as a one-electron carrier instead of ferredoxin (Fd) under iron-limiting conditions
[8]. Meanwhile, the FNR- and Fld-like domains are present in eukaryotic NAD(P)H-dependent enzymes, including cytochrome P450 reductase (cyt P450 reductase)
[9][10] and nitric oxide synthase (NOS) isoforms
[11]. The catalytic domains of these enzymes share a high sequence similarity and structure with FNR and Fld, where the FAD-FMN redox pair donates electrons to final electron acceptors during sequential one-electron transfer reactions
[12]. NADH-cytochrome
b5 reductase (cyt
b5 reductase) shares structural similarities with plant FNR, despite its amino acid sequence similarities being lower
[13]. The cyt
b5 reductase-cyt
b5 complex pair donates an electron to metal-containing proteins
[14]. Cyt P450 reductase (FAD-FMN) and the cyt
b5 reductase (FAD)-cyt
b5 complex exhibit diverse functions in regard to the electron acceptors.
2. Structure and Properties of the FNR, Fd and Fld
The three-dimensional structure of FNR from spinach reveals two subdomains, the NADPH-binding domain and the FAD-binding domain
[1][5]. FNR can transfer electrons to both Fd [2Fe-2S] and Fld (FMN), where an interaction with Fd or Fld occurs in the same structural region of the FAD-binding domain
[1][22]. Both Fd and Fld act as one-electron acceptors or donors, and Fd is replaced with Fld under low-iron conditions
[8][23]. The rate of electron transfer among these proteins is controlled by several factors, including the relative orientation and distance between protein–protein interactions and the differences in the redox potentials between the protein-bound donor and acceptor redox cofactors
[1][3][23][24][25][26]. Sinohara et al.
[1] reported the crystal structures of the
RFNR-
RFd (Fd III) (
R for root) and
LFNR-
LFd (Fd I) (
L for leaf) complexes, and this provides a structural basis for reversing the redox pathway. In the
LFNR-Fd complexes, the distance between the [2Fe-2S] cluster of Fd and the dimethylbenzene edge of the FAD ring is ~5.5–6.0 Å
[1][24], while the
RFNR-Fd complexes can utilize the different sides of the [2Fe-2S] cluster for intermolecular electron transfer. The modeling of the FNR-Fld complexes indicates that the distance between the two flavin rings is ~4.1 Å and that no intervening residues are present between the two cofactors, thus making possible direct one-electron transfer
[25]. Taken together, these observations suggest that the complexes between oxidized FNR and oxidized Fd are relatively stable, as the complexes are stabilized by a salt bridge of FNR33Lys-Fd60Asp between FNR and Fd
[26]. However, this stability is decreased by NADP(H) binding, and unstable complexes can promote the electron transfer rate during the catalytic cycle
[27]. FNR interacts with Fd or Fld in photosynthetic and non-photosynthetic reactions.
The isoalloxazine ring of FAD in the leaf and root FNR is sandwiched between two aromatic residues (Tyr314 and Tyr95), where the phenol ring of carboxyl(C)-terminal Tyr314 shields the face of the isoalloxazine ring of FAD
[5][28]. Thus, C-terminal Tyr314 must move away for productive hydride transfer from reduced FAD to NADP
+, and Ser96 interacts with the N5 atom of the isoalloxazine ring of FAD, where Ser96 forms a hydrogen bond with Glu312. Additionally, C-terminal Tyr314 and the Ser96-Glu312 pair modulates the affinity for NADP
+, the stabilization of FAD semiquinone and the rate of electron transfer
[29]. Therefore, Tyr314 and the Ser96-Glu312 pair are involved in the modulating of the flavin redox properties
[30][31].
The formation of the reduced FAD-NADP
+ (FADH
−-NADP
+) complex with the charge transfer bands of 500–750 nm proceeds via an FAD
ox-NADPH charge-transfer species
[32]. In the presence of excess NADPH (>10-fold), bound NADP
+ is replaced by NADPH: FADH
−-NADP
+ + NADPH
⇌ FADH
−-NADPH + NADP
+, where the FADH
−- NADPH complex does not exhibit significant charge transfer bands. This could be caused by a decrease in π−π stacking interactions between reduced FAD and NADPH. The value of
Eox/red measured using dithionite as a reductant is −377 mV
[32], which is fitted to a Nernstian
n = 2 curve. However, the two-electron reduction potential in the presence of NADP
+ is divided into
Eox/sq (−306 mV) and
Esq/red (−386 mV) (
Table 1), suggesting that the redox potentials of FNR are regulated by the NADP
+/NADPH ratio.
Table 1. Midpoint reduction potentials of FAD and FMN domains.
| Enzyme Specicies |
Eox/sq (mV) |
Esq/red (mV) |
pH |
| FNR (FAD) |
| Spinach (FAD) |
−350 |
−335 |
7 |
| −402 |
−358 |
8 |
| Spinach (FAD) |
−306 |
−386 |
8 |
| (in the presence of NADP+) |
| Anabaena (FAD) |
−385 |
−371 |
8 |
| Anabaena Flavodoxin (FMN) |
−212 |
−436 |
7 |
| Cyt P450 reductase (FAD-FMN) |
| Rabbit (FAD) |
−290 |
−365 |
7 |
| Rabbit (FMN) |
−110 |
−270 |
7 |
| Human (FAD) |
−283 |
−382 |
7 |
| Human (FMN) |
−56 |
−274 |
7 |
| nNOS reductase domain (FAD-FMN) |
| Rat (FAD) |
−260 |
−280 |
7.6 |
| Rat (FMN) |
−50 |
−276 |
7.6 |
| iNOSreductase domain (FAD-FMN) |
| Human (FAD) |
−240 |
−270 |
7 |
| Human (FMN) |
−105 |
−245 |
7 |
The redox potentials for
LFd I and
RFd III are −401 mV and −321 mV, respectively.
[33]. Thus, the one- and two-electron redox potentials of FNR are important factors that control the electron transfer rate (Scheme 1). For the spinach
LFNR, the
Em (FAD/FADH
−) value is −343 mV. Regarding the one-electron redox potential, the oxidized-semiquinone couple (
Eox/sq) is −350 mV, and the semiquinone-fully reduced couple (
Esq/red) is −335 mV at pH 7.0
[34] (
Table 1). Thus, the electron transfer from Fd I to NADP
+ is thermodynamically favorable. On the other hand, the electron transfer from NADPH to Fd III is relatively favorable. In both systems, tissue-specific FNR mediates reversible electron transfer reactions (Equation (1) and Scheme 1)
[33].
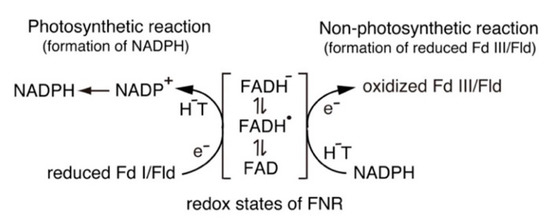
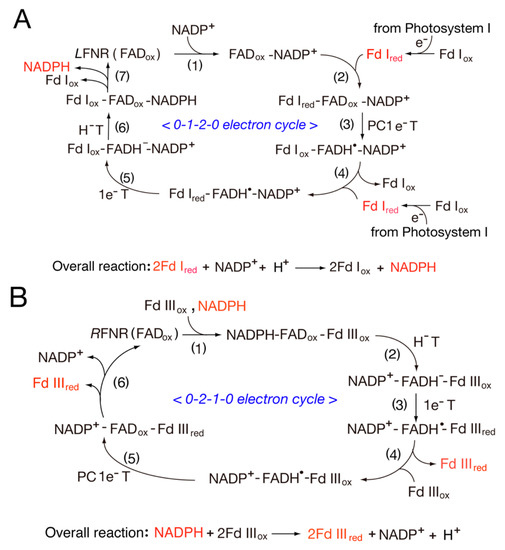
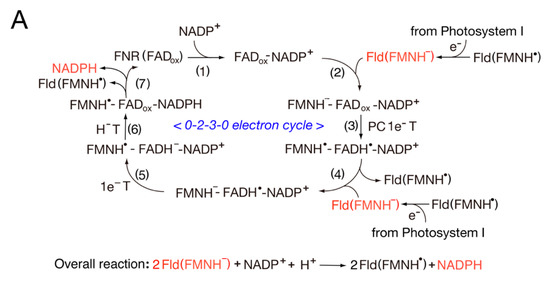
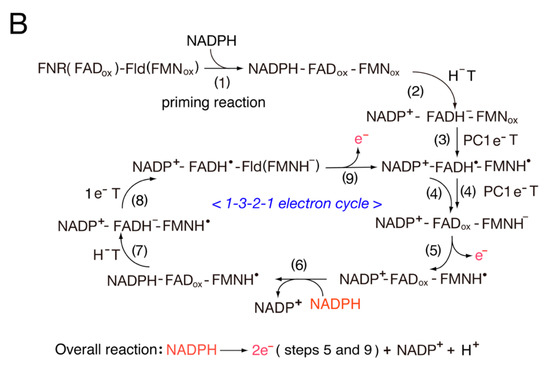
Scheme 1. Switching between one-electron and two-electron transfer reactions in the FNR-Fd I/Fld and FNR-Fd III/Fld systems. H−T, hydride transfer.
Fld is a small electron carrier that participates in low-redox-potential electron transfer pathways, which are classified into three groups based on the presence of tryptophan (Trp) and tyrosine (Tyr) near the isoalloxazine ring of FMN
[35]. In all Flds, the semiquinone formation constant
Ks value is larger than unity, indicating that the
Eox/sq value is always more positive than the
Esq/red value
[36]. The
Eox/sq value is associated with the proton-coupled one-electron reduction from FMN to FMNH
•, while
Esq/red is associated with the one-electron reduction from FMNH
• to FMNH
−. The
Eox/sq value is shifted from ~−240 mV to less-than-negative values, while
Esq/red is shifted from ~−170 mV to ~−540 mV, depending on the pH and the species of Fld. The neutral semiquinone, FMNH
•, is stabilized by the hydrogen bond of the carbonyl and amide groups of the protein backbone with the N5 atom of the isoalloxazine ring of the flavins. Its reactivity is lower than that of the fully reduced form, where the FMNH
•/FMNH
− couple acts as a one-electron carrier
[36].
Flds possess a unique structure, and the loop structures in the FMN environment play an important role in electron transfer reactions
[37]. The FMN binding sites in the bacterial Fld and eukaryotic diflavin reductase family possess similar loop structures and are sandwiched between two aromatic amino acid residues. Rwere et al.
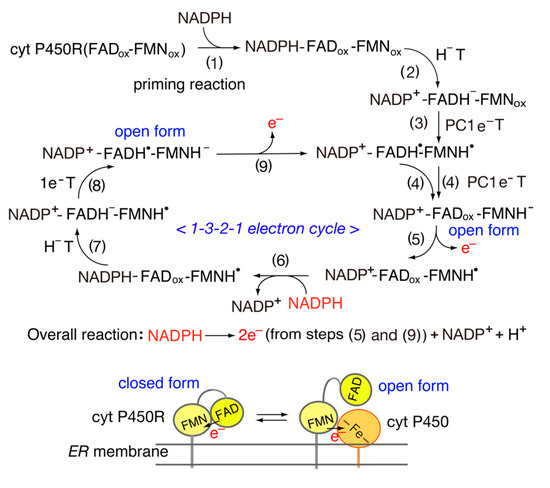
[38] reported that the length and sequence of the “140 s” FMN binding loop of cyt P450 reductase functions as a key determinant of its redox potential and activity with cyt P450s. The one-electron redox potentials of cyt P450 reductase (FAD-FMN)
[39][40] and the reductase domain (FAD-FMN) of NOS isoforms
[41][42] are different from those of plant FNR (FAD)
[34] and
Anabaena flavodoxin (FMN)
[43], but the one-electron redox potentials of FAD and FMN always satisfy
Eox/sq >
Esq/red (Ref.
[14] and
Table 1).
The kinetic parameters for Fd reduction by FNR using the NADPH-dependent cyt
c reductase assay (NADPH → FNR → Fd/Fld → cyt
c) were reported
[29]. The values for pea FNR are in the range of those of the
Anabaena enzyme, with a slightly larger
kcat and a smaller
Km (
Table 2)
[29], while Kimata-Ariga and co-workers recently reported the tissue-specific parameters, where maize
RFNR has a higher
Km value for Fd I than for Fd III
[1]. In addition, the orientation of the
RFNR-Fd complex remarkably varies from that of the
LFNR-Fd complex
[1], suggesting that the root and leaf complexes utilize the different sides of the [2Fe-2S] cluster for the intermolecular electron transfer, which might lead to the evolutional switch between photosynthetic and heterotrophic assimilation.
Table 2. Kinetic parameter of Pea and Anabaena FNR in NADPH-dependent cytochrome c reductase activity.
| FNR Species |
KFdcat (s−1) |
KFdm (µM) |
KFdcat/KFdm (µM s−1) |
KFldcat (s−1) |
KFldm (µM) |
KFldcat/KFldm (µM s−1) |
| Pea FNR |
139 |
6.5 |
21.3 |
30.6 |
16.7 |
1.8 |
| Anabaena FNR |
200 |
11 |
18.2 |
23.3 |
33 |
0.7 |