1. Introduction
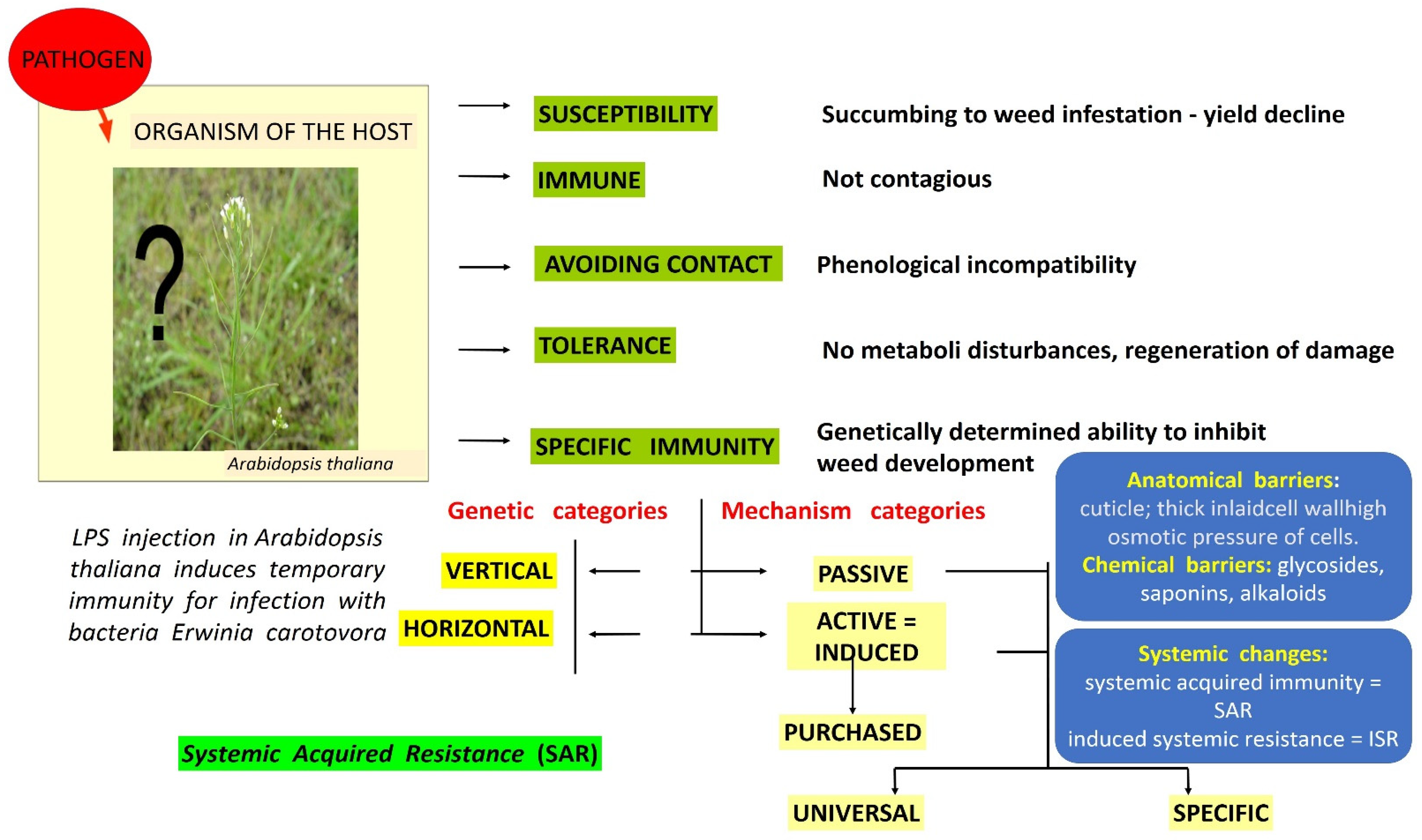
Plants have excellent strategies that use many constitutive or induced anatomical, molecular and biochemical defense mechanisms (Figure 1).
Figure 1. Plant resistance to biotic and abiotic factors. Source: own.
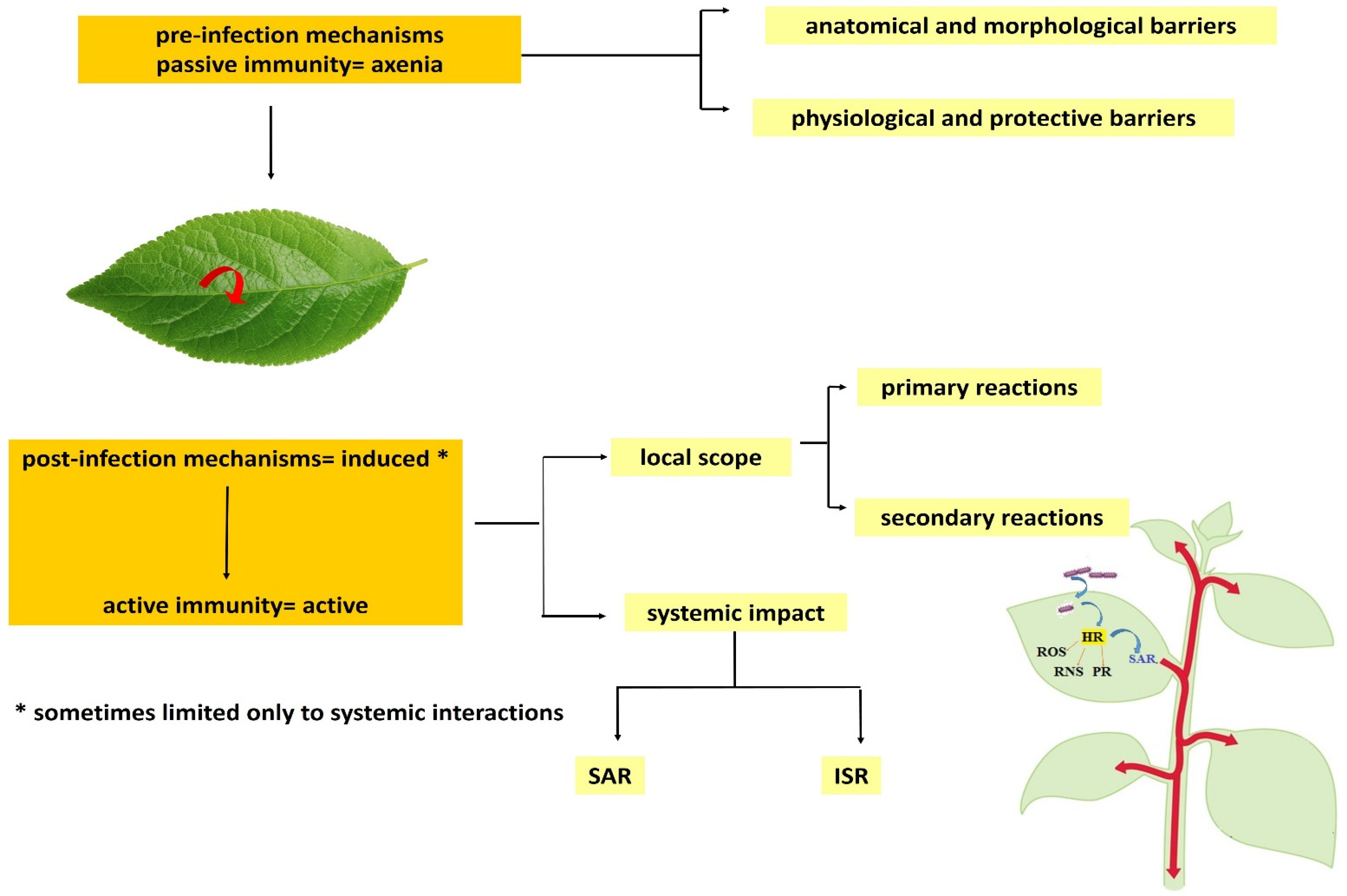
Induced immune responses are triggered in response to a direct, sudden attack by a pathogen, injury or contact with a trigger molecule, or elicitor. They are a signal for the mobilization of the defense system in the whole plant, and the appearance of so-called systemic resistance. There is systemic induced resistance (ISR—induced system resistance) that is triggered by contact with non-pathogenic organisms or synthetic elicitors, and systemic acquired resistance (SAR—system acquired resistance), as a result of prior contact with the pathogen. However, as a result of the emergence of new data, this division is being systematically modified. Higher plants do not have specialized, mobile immune cells or antibodies; however, they are able to effectively defend themselves against the attack of pathogens, since in the case of plants, almost every cell triggers an effective defense response as a result of the effective functioning of the immune system. Constitutive defense mechanisms protect the entire plant passively, through the following: physical barriers, such as the cell wall and the cytoskeleton; waxes (composition) on the surface of the plant; and secondary metabolites that are produced constitutively, e.g., saponins (Figure 2).
Figure 2. Induced and active immunity. Source: own.
One of the models that describes the immune system of plants is the zigzag model presented by Jones and Dangle, which concerns two levels of resistance. Induced defense mechanisms defend the plant against both host and non-host pathogens, and are induced by the appearance of attendant signals that are related to the presence of pathogenic organism-elicitors
[1][2]. This model was the first to demonstrate the complex interplay between plants and their pathogens, and is extremely useful.
Jons and Dangle’s
[2] zigzag model shows the successive steps in plant-pathogen interaction. First, it presents pathogen-related molecular patterns (PAMPs). They are recognized by plant transmembrane PRR receptors, which allow for the emergence of molecular pattern-induced immunity (PTI); finally, the activation of a defense reaction occurs. In the next phase, the infectious pathogen provides effectors that either interfere with PTI, or enhance the nutrition and spread of the pathogen, ultimately leading to the development of sensitivity (ETS). Only in the third phase of this process can effectors become avirulence factors (avr), when the plant already has the appropriate resistance proteins (R). Recognition of the immune effector by the R protein activates a specific immune effector trigger (ETI), which leads to the development of a hypersensitivity reaction (HR). In the next phase, the pathogen may gain a new effector, e.g., in horizontal gene transfer, which may reduce an ETI. In response, selection may favor new plant R alleles whose products recognize the new effector, again leading to the activation of the ETI
[1][2].
2. Resistance of Weeds to Herbicides
Weed herbicide resistance is arguably the most important research area in the field of weed science, which results from the excessive and one-way use of active substances in herbicides. As a consequence, individuals that contain resistance genes are selected from the weed population, which results in the depletion of the genetic pool of agro-ecosystems, and in an increase in the costs of eliminating resistant weeds
[3][4].
The use of herbicides in arable crops triggers the phenomenon of selection. Susceptible plants are killed, while herbicide-resistant plants can survive and reproduce without competition from herbicide-sensitive plants
[5]. If the herbicide is used continuously, resistant plants successfully reproduce and become dominant in the population. After the herbicides enter the plant, they are transported to the site of activity. This transport can take place both in inanimate (apoplast, e.g., cell walls, xylem) or living tissues (symplast, e.g., cellular plasmalemma)
[5][6]. The movement of herbicides can take place over short distances (activity in living cells close to entry), as well as over longer distances, through the vascular system of plants
[7][8].
Herbicide resistance may result from several mechanisms, among which loss of target site sensitivity appears to be the most important. Weeds may be resistant to one specific herbicide, which is called simple or single resistance. Resistant biotypes may also have a mixed (cross) resistance to at least two herbicides, with the same mechanism of action, but with different chemical structures. However, our knowledge of specific DNA changes that confer resistance at non-target sites is still in the early stages
[3][9]. Currently, there is also so-called multiple resistance, consisting of the insensitivity of a specific weed biotype to at least two herbicides from different chemical groups, and with different mechanisms of action.
Numerous resistance-conferring mutations in weeds have been identified from dozens of weed species, and now include nine herbicide target sites in chronological order, such as the following: D1 protein (atrazine), acetolactate synthase (chlorimuron), tubulin (trifluralin), acetyl CoA carboxylase (clethodim), 5-enolypyruvylshikimate-3-phosphate synthase (glyphosate), phytoene desaturase (fluridone), protoporphyrinogen, oxidase (glufosinate), and auxin receptor (2,4-D)
[3][5][6][9][10][11]. Beckie
[3] has provided the most recent update of the new mutations that have recently been identified for each of these nine targets. He believes that “new mutations” are those that trigger an amino acid change that has not been previously reported in any of the weed species. Although it has already been observed that gene duplication events confer resistance to groups 1 and 9 herbicides, the basic genetic mechanisms of resistance evolution lie outside the corresponding target coding region.
The International Herbicide-Resistant Weed Database
[12] presents the numbers of species that are resistant to each site of action. As many species have developed resistance to more than one site of action, the sum total represents the unique cases of resistance, not the number of species. Thus far, over 514 species of weeds that are resistant to various active substances have been identified. Of these, 268 are dicotyledons, and 246 are monocotyledons. In the world, the greatest number of biotypes (as many as 170) show resistance to acetolactate synthetase (ALS) inhibitors. Herbicide-resistant weeds have been found in ninety-three crops in seventy-two countries
[12][13][14][15][16]. There are biotypes of weeds that are known to have naturally developed resistance to triazine herbicides, by mutating the D1 protein gene
[16]. This resistance can also be due to the rapid degradation or conjugation of herbicides. Glutathione-S-transferase and cytochrome P450 monooxygenase are the main enzymes that are involved in these processes
[4][17].
In nature, we can encounter cross resistance, which occurs when there is resistance to two or more herbicides, with the same or different mode of action, resulting from the presence of a single immune mechanism (one genetic mutation). The mechanism of weed resistance to herbicides from the group of ALS inhibitors (according to the HRAC classification marked with the letter B) has been very well described
[12][16][17][18]. The resistance of these substances to herbicides is most often associated with the Pro197 mutation. As a result, the amino acid proline, at position 197, can be replaced with a different group of amino acids. The most common, however, is the Pro197-Ser mutation in weed-resistant biotypes. Many researchers
[16][19][20][21][22] proved that using sulfometron to control different biotypes of weeds that are resistant to ALS herbicides can be strictly determined, whether it is either mutational or non-mutational resistance.
Another type of resistance is multiple resistance, which is a resistance to herbicides from more than one chemical class to which a given population has been exposed
[23]. It refers to a biotype of a weed or crop that has developed resistance mechanisms to more than one herbicide. This resistance arose as a result of separate selection processes. Multiple resistance has been reported for weed species such as the following:
L. rigidum,
A. myosuroides,
C. canadensis,
A. palmeri and
A. tuberculatus [24][25][26][27]; in the case of spring barley and wheat, with active ingredients such as chlorpropham, chlorsulfuron, clomazone, diclofopmethyl, ethalfluralin, fluazifop butyl, imazapyr, metolachlor, metsulfuron-methyl, quisalophop-ethyl, sethoxydim, tralkoxydim, triallate, triasulfuron and trifluralin); multiple resistance was found in South Australia in as many as 6 sites of action: inhibition of acetyl-CoA carboxylase HRAC group 1 (Older version A), group 2 (older version B), inhibition of microtubule formation HRAC group 13 (older version F4); inhibition of microtubule formation 2HRAC group 3 (older K1); inhibition of microtubule organization HRAC group 23 (older version of K2); and inhibitors of the synthesis of very long-chain fatty acids HRAC group 15 (older version of K3 N)
[12].
Synthetic auxins (HRAC O group) one, which mimic the naturally occurring plant hormone indole-3-acetic acid (IAA), have a wide range of use in the selective control of broadleaf weeds in grass crops. SAHs are divided into several subclasses which include the following: (1) phenoxycarboxylates, (2) benzoates, (3) pyridine carboxylates, (4) pyridyloxycarboxylates, (5) quinolone carboxylates, (6) pyrimidine carboxylates and (7) aryl picolinates. Each subclass has a distinct chemical structure. SAHs have been used since the introduction of 2,4-D (1945) until now, with the introduction of florpiraxifene-benzyl in 2018. In order to maintain the usefulness of SAH, it is essential to gain more knowledge about resistance mechanisms, and how selection and subsequent evolution have taken place in weed species that are resistant to SAH
[26].
Mechanisms of synthetic resistance to auxin herbicides is well-known in weed species.
Papaver rhoeas is the most common dicotyledonous weed species in winter cereals in Europe. This cross-pollinated species is difficult to control due to its high seed production, persistent seed banks and extended seed germination times. With the emergence and spread of herbicides, the resistance of
P. rhoeas made it become an increasingly nuisance weed, especially in southern Europe. Phenoxy-carboxylate (2,4-D) and biotypes of this weed species that are resistant to MCPA and ALS inhibitors (tribenuron methyl) have been reported for 10 years in Spain, France and Greece. Few studies have been performed to reveal the mechanisms and genes that are involved in
P. rhoeas resistance to SAH. The lack of 2,4-D translocation in resistant plants may contribute to their immune response. The production of ethylene in sensitive- and 2,4-D-treated plants was 4–8 times higher than in resistant plants
[26]. According to Beckie
[3], 2,4-D may not reach the nuclear protein receptor complex in resistant plants, which in turn causes the repression of auxin genes, some of which are responsible for ethylene production. Accumulation of ethylene in
P. rhoeas cells can inhibit photosynthesis and produce H
2O
2, a reactive oxygen species, which leads to plant death.
In 2017–2020, the ConResi consortium that consisted of scientific institutes, agricultural and natural science universities and pharmaceutical companies, implemented a project on the following subject: “Strategy for counteracting weed resistance to herbicides as an important factor in ensuring sustainable development of the agroecosystem”. The aim was to develop and implement a holistic strategy to reduce the risk of spreading herbicide-resistant weed biotypes, and to develop means to control them. Four species of weeds were investigated: (1) field foxtail (
Alopecurus myosuroides Huds.), (2) grain broom (
Apera spica-venti (L.) P. Beauv), (3) field poppy (
Papaver rhoeas L.) and (4) cornflower (
Centaurea cyanus L.). In total, over 2150 samples of seeds of the above-mentioned species of weeds from all over Poland were assessed, and were subjected to biological tests for sensitivity to the following herbicides: 2,4-D, chlortoluron, dicamba, fenoxaprop-P-ethyl, florasulam, iodosulfuron, pendimethalin, pinoxaden, piroxulam and tribenuron-methyl
[1][28]. Resistant biotypes were found in each of the tested species, whereas monocotyledonous weeds, i.e., field grasshopper and grain broom, turned out to be more resistant to herbicides than dicotyledonous weeds (field poppy and cornflower). Herbicide-resistant biotypes were subjected to molecular analyses, in order to determine the genetic basis of their resistance. Moreover, the environmental adaptations of resistant biotypes, as well as their competitiveness with winter wheat, were analyzed in a series of pot experiments that were carried out at various experimental stations and laboratories in Poland
[29]. Synowiec et al.
[30] demonstrated that the competitive effect of winter wheat on herbicide-susceptible or resistant bractbill biotypes is specific to local conditions. Under drought conditions, the biotype of bumblebee with multiple herbicide resistance turned out to be more competitive with winter wheat. In addition, the project included activities that enabled the implementation of research results into agricultural practice, particularly their dissemination among farmers
[4][31].
An integral element of herbicide resistance management is the task of many authors
[3][12][16][20][26] to periodically analyze the sensitivity of the population of weed species in the agro-region to commonly used herbicides. The analyses should provide information on inter- and intra-population variability in the effective dose (ED) required for 50 or 90%, as well as the reduction in biomass viability. Hence, an analysis of weed sensitivity to herbicides will help determine whether populations become less sensitive to the herbicide over time, and whether labeling rates need to be adjusted. Such studies are important in mitigating the quantitative (creeping) evolution of resistance, especially for key herbicides, such as glyphosate, and major problematic crossing weeds such as Lolium spp.
[5][32]. Sublethal doses of the herbicide may even alter the metabolism, growth and survival of susceptible plant species that are highly selfish, such as
Avena fatua L. (wild oats)
[6].
Natural selection for herbicide-resistant weed genotypes can affect genetic variability or the genetic and physiological background that is variable, due to stress responses from exposure to sublethal herbicides. Stress-induced changes concern DNA mutations, epigenetic changes, transcription and protein modification, all of which can lead to herbicide resistance and pleiotropic effects
[3][9]. The results of these studies are being developed, and provide great hope for their implementation, not only in Poland, but also in Europe. In light of the current status of weed resistance to herbicides in Poland, as well as the implementation of the Green Deal Directive in the European Union, research on this phenomenon is gaining importance, especially regarding the impact of biotypes that are resistant to the functioning of agrocenoses and natural biocenoses, and their economic impact. Adamczewski et al.
[15] conducted research on weed resistance to herbicides in Poland. About 50% of the analyzed samples showed resistance to sulfonylurea herbicides. Resistance to acetyl-CoA carboxylase (ACCase) inhibitors was found in eighteen fields, and resistance to PSII inhibitors (isoproturon) was found in twelve fields. The resistance of
Alopecurus myosuroides Huds. to herbicides was recorded in as many as twenty-six tested fields. In ten spring crops, they found that
Avena fatua L. was resistant to acetyl-CoA carboxylase inhibitors. The resistance of
Centaurea cyanus L. to tribenuron methyl was confirmed in twenty-three fields.
Matricaria inodora L. and
Papaver rhoeas L. proved to be resistant to methyl volcano in several fields of winter wheat. Moreover, three biotypes of
Chenopodium album L. and two biotypes of
Amaranthus retroflexus L. were resistant to metamitron.
Conyza canadensis L.,
Lolium spp. showed resistance to glyphosate
[14][33]. Empirical observations by Panozzo et al.
[33] generally showed that there was a shift and decrease in susceptibility of Lolium spp. to glyphosate in Italy. This is likely due to the long-term use of glyphosate, and to the sublethal doses used. Therefore, it is necessary to determine the variability of response of
Lolium spp. to glyphosate, and to identify the optimum field dose.
3. Resistance of Plants to Pesticides
Pesticide resistance is defined as the change in the sensitivity of a pest population to a pesticide, resulting in failure to control the pest when the pesticide is properly applied. Resistance can develop whenever the same or a similar pesticide with the same mode of action is used repeatedly. It is also believed that pests change or mutate to become resistant. However, it is not a single pest (e.g., an insect, pathogen, or micro-organism) that changes, but the entire population which changes. When a pesticide is applied to a crop or treatment site, only a small fraction of the pest’s population (e.g., one insect, one crop, or one in ten million weeds) can survive exposure to the pesticide, due to its genetic makeup. When surviving pests reproduce, some of their young inherit a genetic trait that confers resistance to the pesticide. These pests will not be damaged the next time a similar pesticide is used. If the same pesticide is used frequently, the percentage of the less susceptible will increase in the population. When a pesticide applicator recognizes that a once highly effective pesticide is not limiting or destroying the pest at the same rate, higher doses and more frequent applications become necessary, until a point is reached when the pesticide provides little or no control. Then, it can be considered that the population has become resistant
[3][5]. With global warming and structural changes in agricultural production, epidemics of crop diseases are becoming more frequent, and it is possible that new types or variants of phytopathogens will emerge and evolve. As a result, the health of cultivated plants is under threat, and plant production is facing more frequent challenges. However, new methods for identifying and characterizing pathogens, in addition to better understanding of the mechanisms that govern plant-microbial interaction are critical to designing crop health management strategies, in order to minimize pathogen-related crop losses. Over the last two decades, tremendous progress has been made in deciphering the molecular mechanisms of plant resistance using model patho-systems. However, the disease resistance mechanisms of many crops remain poorly characterized, due to the genetic/genomic complexity of many crop species, and/or a poor understanding of their pathogens. However, next-generation DNA sequencing technologies have enabled the sequencing and characterization of the genomes of many crop species and their pathogens, which in recent times has greatly facilitated research into the mechanisms of crop–pathogen interactions. Such new knowledge should help to improve crop health and/or reduce pesticide use, and thus contribute to sustainable agriculture
[3][5][8][9][13][14][34].
The pesticide detoxification system involves a three-phase process: Phase I—pesticide activation through hydrolysis, oxidation or reduction, induced by enzymes such as carboxylesterase, cytochrome P450 monooxygenases and peroxidase. Phase II—the activated pesticide is coupled with amino acids, glucose or glutathione; conjugation is catalyzed by the enzymes glutathione S-transferase and UDP-glycosyltransferase. In phase III, the conjugated pesticides are transferred to vacuoles/apoplasts, or bind to the cell wall/lignin
[5][13][35]. In agricultural crops, the most common mode of herbicide tolerance is metabolism induced by changes by enzyme systems, such as cytochrome P450 (CYP) monooxygenases; glutathione S-transferases (GSTs); and glucosyl transferases (GTs). These enzymes, as well as cofactors such as reduced glutathione (GSH), are activated by certain chemicals called safeners
[14]. Safeners are, in turn, used in conjunction with herbicides to tolerate crops such as
Triticum aestivum L.,
Oryza sativa L. and
Zea mays L.
[14][15].
Immunity inducing factors are elicitors, i.e., compounds released when interacting with a potential pathogen, which, when recognized by the appropriate receptors, trigger a cascade of events that leads to the generation of plant resistance. Elicitors induce a defense response in the plant, through receptors that transmit a signal to the cell nucleus. It is a group of very diverse proteins that are synthesized by plants. Due to the similarities in their domain structure, they were divided into five classes. R proteins interact with the avirulence factors Avr, secreted by bacteria, fungi, and the Avr proteins of viruses. Most of the currently known R proteins are found in the cytosol, suggesting that they recognize intracellular Avr factors. The following was found in the genome of the model
Arabidopsis plant: 149 genes that encode R proteins of the NB-LRR type, including the only known function-participation in defense reactions; 233 genes that code for LRK proteins; 110 genes that code for proteins of the RLP type
[3][16]. Due to the type of triggered immunity, there are common elicitors and race-specific elicitors. Common elicitors (i.e., β-glucans, chitin, polypeptides, glycolipids) are produced by many pathogens, and interact with all cultivars of the affected host plant species. They are responsible for the liberation of so-called racial-non-specific resistance (field, horizontal, partial). Race-specific elicitors (products of avr avirulence genes) are active only in variants that have a complementary resistance gene (R). They are the prerequisite for the occurrence of type resistance gen-na-gen, or race-specific (vertical, full) resistance. Rapid reactions that occur mainly in the periphery of the cell, which do not require gene expression, are a direct consequence of recognition of a pathogen/elicitor by a plant cell. They include the following: membrane depolarization, ion flux across the membrane (K
+ and Cl
− outflow and H
+ and Ca
2+ inflow), and oxygen explosion. The ‘gene to gene’ hypothesis assumes that the recognition of the pathogen, and the activation of defense reactions, occurs only when the plant possesses the appropriate R resistance gene, and is attacked by micro-organisms that carry the corresponding avr avirulence gene. The pathogen with the avr gene is pathogenic to plants that do not have the appropriate R gene-no-defense response
[3]. The main precursor in the synthesis of SA is phenylalanine. In the first stage, this amino acid is converted into trans-cinnamic acid with anti-auxin properties, by means of the PAL enzyme (ammonia-L-phenylalanine), in the process of non-oxidative deamination. This acid, depending on environmental conditions and under the influence of a specific isomerase, can be transformed into cis-cinnamic acid, which has auxin-like properties. Trans-cinnamic acid in the oxidation process turns into benzoic acid, which then undergoes hydroxylation to become salicylic acid. In turn, the hypothetical pathway leads to the transformation of trans-cinnamic acid into trans-coumaric acid, which is converted to cis-coumaric acid, which forms the basic coumarin skeleton
[16]. The markers of the resistance reaction are phenylalanine ammoniaolase (PAL); chalcone isomerase (CHI); isoflavone reductase (IFR); cinnamate 4-hydroxylase; 4-coumaryl-CoA ligase; chalcone synthase (CHS); prenylotransferase
[5]; pathogenesis-related proteins (PR), and pathogen-dependent stress associated with pathogenesis. The components of the signal chain in the immune reaction are PR proteins, which are mainly hydrolytic enzymes: β- (1,3) endoglucanases and chitinases, as well as peroxidases. The first to describe the PR of tobacco infected with mosaic virus was Ivanoski, who reported in 1892 that extracts from contaminated leaves were still contagious when filtered through a Chamberland filter candle. However, Ivanowski probably did not fully understand the meaning of his discovery. Beijerinck, in 1898, was the first to name "virus", the initiator of the tobacco mosaic. Thus, Ivanovski and Beijerinck made an unequal but decisive and complementary contribution to the discovery of viruses. The functions of PR proteins include creating a protective barrier against pathogens by separating them and collecting them at the site of infection; weakening the sensitivity of plants; showing antibacterial and antifungal activity in vitro (some antiviral). They are expressed as the result of a hypersensitivity reaction. Their expression depends on the intensity and type of attack. They can mediate plant resistance to the pathogen by overexpressing their genes in transgenic plants. The prevalence of antifungal proteins correlates with the prevalence of fungi that colonize plants, animals and humans (in the last decade there has been a drastic increase in fungal infections—about two hundred human fungal pathogens)
[8][13][16][34].
When plants are infected, both plants and pathogens secrete protein-containing molecules that determine their interactions. While proteins secreted by the pathogen are involved in infection and pathogenicity, proteins secreted by plants play a key role in their resistance. Host–pathogen interaction is defined as the way in which microbes or viruses persist in the host organism at the molecular level, and then the cellular level, the organism level, and finally at the population level. The term is most often used to refer to pathogenic micro-organisms, although they may not cause disease in all intermediate hosts
[16].