
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dean Liu | -- | 1230 | 2022-11-09 01:39:33 |
Video Upload Options
Influenza hemagglutinin (HA) or haemagglutinin[p] (British English) is a glycoprotein found on the surface of influenza viruses. Being a class I fusion protein, it is responsible for binding the virus to cells with sialic acid on the membranes, such as cells in the upper respiratory tract or erythrocytes. It is also responsible for the fusion of the viral envelope with the endosome membrane, after the pH has been reduced. The name "hemagglutinin" comes from the protein's ability to cause red blood cells (erythrocytes) to clump together ("agglutinate") in vitro.
1. Subtypes
Influenza A can be categorized based on the different subtype of proteins present: Hemagglutinin (HA) and Neuraminidase (NA).[1] HA has at least 18 different subtypes. These subtypes are named H1 through H18. H16 was discovered in 2004 on influenza A viruses isolated from black-headed gulls from Sweden and Norway . H17 was discovered in 2012 in fruit bats.[2][3] Most recently, H18 was discovered in a Peruvian bat in 2013.[4] The first three hemagglutinins, H1, H2, and H3, are found in human influenza viruses. Neuraminidase (NA) has 11 known subtypes, hence influenza virus is named as H1N1, H5N2 etc., depending on the combinations of HA and NA.
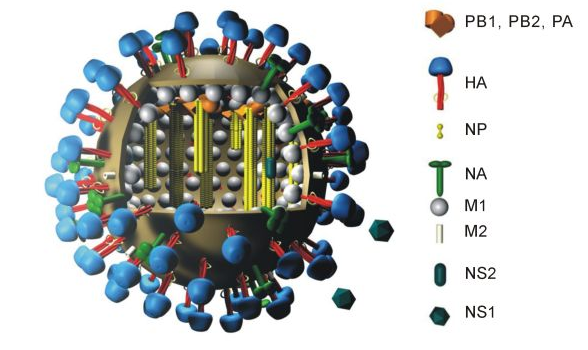
A highly pathogenic avian flu virus of H5N1 type has been found to infect humans at a low rate. It has been reported that single amino acid changes in this avian virus strain's type H5 hemagglutinin have been found in human patients that "can significantly alter receptor specificity of avian H5N1 viruses, providing them with an ability to bind to receptors optimal for human influenza viruses".[5][6] This finding seems to explain how an H5N1 virus that normally does not infect humans can mutate and become able to efficiently infect human cells. The hemagglutinin of the H5N1 virus has been associated with the high pathogenicity of this flu virus strain, apparently due to its ease of conversion to an active form by proteolysis.[7][8]
2. Structure
General Structure
- HA is a homotrimeric integral membrane glycoprotein. It is shaped like a cylinder, and is approximately 13.5 nanometres long.[9][10] HA trimer is made of three identical monomers. Each monomer is made of an intact HA0 single polypeptide chain with HA1 and HA2 regions that are linked by 2 disulfide bridge.[11][12] Each HA2 region adopts alpha helical coiled coil structure and sits on top of the HA1 region, which is a small globular domain that consists of a mix of α/β structures.[13] The HA trimer is synthesized as inactive precursor protein HA0 to prevent any premature and unwanted fusion activity and must be cleaved by host proteases in order to be infectious. At neutral pH, the 23 residues near the N-terminus of HA2, also known as the fusion peptide that is eventually responsible for fusion between viral and host membrane, is hidden in a hydrophobic pocket between the HA2 trimeric interface.[14] The C-terminus of HA2, also known as the transmembrane domain, spans the viral membrane and anchors protein to the membrane.[15]
HA1
- HA1 is mostly composed of antiparallel beta-sheets.[16]
HA2
- HA2 domain contains three long alpha helices, one from each monomer. Each of these helices is connected by a flexible, loop region called Loop-B (residue 59 to 76).[17]
3. Function of HA in Viral Entry
HA plays two key functions in viral entry. Firstly, it allows the recognition of target vertebrate cells, accomplished through the binding to these cells' sialic acid-containing receptors. Secondly, once bound it facilitates the entry of the viral genome into the target cells by causing the fusion of host endosomal membrane with the viral membrane.[18]
Specifically, HA1 domain of the protein binds to the monosaccharide sialic acid which is present on the surface of its target cells, allowing attachment of viral particle to the host cell surface. The host cell membrane then engulfs the virus, a process known as endocytosis, and pinches off to form a new membrane-bound compartment within the cell called an endosome. The cell then attempts to begin digesting the contents of the endosome by acidifying its interior and transforming it into a lysosome. Once the pH within the endosome drops to about 5.0 to 6.0, a series of conformational rearrangement occurs to the protein. First, fusion peptide is released from the hydrophobic pocket and HA1 is dissociated from HA2 domain. HA2 domain then undergoes extensive conformation change that eventually bring the two membranes into close contact.
This so-called "fusion peptide" that was released as pH is lowered, acts like a molecular grappling hook by inserting itself into the endosomal membrane and locking on. Then, HA2 refolds into a new structure (which is more stable at the lower pH), it "retracts the grappling hook" and pulls the endosomal membrane right up next to the virus particle's own membrane, causing the two to fuse together. Once this has happened, the contents of the virus such as viral RNA are released in the host cell's cytoplasm and then transported to the host cell nucleus for replication.[19]
- Monoclonal Antibodies/ Broadly Neutralizing Antibodies
Since hemagglutinin is the major surface protein of the influenza A virus and is essential to the entry process, it is the primary target of neutralizing antibodies. Neutralizing antibodies against flu have been found to act by two different mechanisms, mirroring the dual functions of hemagglutinin:
- Approach 1: Inhibition of attachment to target cells
- Some antibodies against hemagglutinin act by inhibiting attachment. This is because these antibodies bind near the top of the hemagglutinin "head" (blue region in figure above) and physically block the interaction with sialic acid receptors on target cells.[20]
- Approach 2: Inhibition of membrane fusion between host cell and virus
- This group of antibodies acts by preventing membrane fusion (only in vitro; the efficacy of these antibodies in vivo is believed to be a result of antibody-dependent cell-mediated cytotoxicity and the complement system).[21]
- The stem or stalk region of HA (HA2), is highly conserved across different strains of influenza viruses.[22] Its structural changes from prefusion to postfusion conformation drives fusion between viral membrane and host membrane. Therefore, Antibodies targeting this region can block key structural changes that eventually drive the membrane fusion process, and therefore are able to achieve antiviral activity against several influenza virus subtypes. At least one fusion-inhibiting antibody was found to bind closer to the top of hemagglutinin, and is thought to work by cross-linking the heads together, the opening of which is thought to be the first step in the membrane fusion process.[23]
- Examples are human antibodies F10,[24] FI6,[25] CR6261. They recognize sites in the stem/stalk region (orange region in figure at right), far away from the receptor binding site.[26][27]
- In 2015 researchers designed an immunogen mimicking the HA stem, specifically the area where the antibody ties to the virus of the antibody CR9114. Rodent and nonhuman primate models given the immunogen produced antibodies that could bind with HAs in many influenza subtypes, including H5N1.[28] When the HA head is present, the immune system does not make bNAbs (broadly neutralizing antibodies). Without the head, the whole protein becomes unrecognizable to antibodies. One team designed self-assembling HA-stem nanoparticles, using a protein called ferritin to hold the HA together. Another replaced and added amino acids to stabilize a mini-HA lacking a proper head.
- Other hemagglutinin-targeted influenza virus inhibitors[29] that are not antibodies:
- Arbidol
- Small Molecules
- Natural compounds
- Proteins and peptides
References
- "Influenza Type A Viruses | Avian Influenza (Flu)" (in en-us). 2017-04-19. https://www.cdc.gov/flu/avianflu/influenza-a-virus-subtypes.htm.
- "Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls". Journal of Virology 79 (5): 2814–22. March 2005. doi:10.1128/JVI.79.5.2814-2822.2005. PMID 15709000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=548452
- Unique new flu virus found in bats http://www.nhs.uk/news/2012/03march/Pages/cdc-finds-h17-bat-influenza.aspx
- "New world bats harbor diverse influenza A viruses". PLoS Pathogens 9 (10): e1003657. October 2013. doi:10.1371/journal.ppat.1003657. PMID 24130481. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3794996
- "Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses". Biological & Pharmaceutical Bulletin 28 (3): 399–408. March 2005. doi:10.1248/bpb.28.399. PMID 15744059. https://dx.doi.org/10.1248%2Fbpb.28.399
- "Evolution of the receptor binding phenotype of influenza A (H5) viruses". Virology 344 (2): 432–8. January 2006. doi:10.1016/j.virol.2005.08.035. PMID 16226289. https://dx.doi.org/10.1016%2Fj.virol.2005.08.035
- "Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses". Science 293 (5536): 1840–2. September 2001. doi:10.1126/science.1062882. PMID 11546875. https://dx.doi.org/10.1126%2Fscience.1062882
- "Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential". Avian Diseases 40 (2): 425–37. 1996. doi:10.2307/1592241. PMID 8790895. https://dx.doi.org/10.2307%2F1592241
- "Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution". Nature 289 (5796): 366–73. January 1981. doi:10.1038/289366a0. PMID 7464906. https://dx.doi.org/10.1038%2F289366a0
- "Hemagglutinin-Mediated Membrane Fusion: A Biophysical Perspective". Annual Review of Biophysics 47 (1): 153–173. May 2018. doi:10.1146/annurev-biophys-070317-033018. PMID 29494252. https://dx.doi.org/10.1146%2Fannurev-biophys-070317-033018
- "Hemagglutinin-Mediated Membrane Fusion: A Biophysical Perspective". Annual Review of Biophysics 47 (1): 153–173. May 2018. doi:10.1146/annurev-biophys-070317-033018. PMID 29494252. https://dx.doi.org/10.1146%2Fannurev-biophys-070317-033018
- "Modulation of the pH Stability of Influenza Virus Hemagglutinin: A Host Cell Adaptation Strategy". Biophysical Journal 110 (11): 2293–2301. June 2016. doi:10.1016/j.bpj.2016.04.035. PMID 27276248. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=4906160
- Smrt, Sean T.; Lorieau, Justin L. (2016), "Membrane Fusion and Infection of the Influenza Hemagglutinin" (in en), Advances in Experimental Medicine and Biology (Springer Singapore): pp. 37–54, doi:10.1007/5584_2016_174, ISBN 9789811069215 https://dx.doi.org/10.1007%2F5584_2016_174
- "The structure and function of the hemagglutinin membrane glycoprotein of influenza virus". Annual Review of Biochemistry 56 (1): 365–94. June 1987. doi:10.1146/annurev.bi.56.070187.002053. PMID 3304138. https://dx.doi.org/10.1146%2Fannurev.bi.56.070187.002053
- H., Strauss, James (2008). Viruses and human disease. Strauss, Ellen G. (2nd ed.). Amsterdam: Elsevier / Academic Press. ISBN 9780080553160. OCLC 630107686. https://www.worldcat.org/oclc/630107686.
- "Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution". Nature 289 (5796): 366–73. January 1981. PMID 7464906. http://www.ncbi.nlm.nih.gov/pubmed/7464906
- "Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus". Science 303 (5665): 1866–70. March 2004. doi:10.1126/science.1093373. PMID 14764887. https://dx.doi.org/10.1126%2Fscience.1093373
- "Attachment and entry of influenza virus into host cells. Pivotal roles of hemagglutinin". Structural Biology of Viruses. Oxford University Press. 1997. pp. 80–104.
- "Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection". Biochimica et Biophysica Acta 1838 (4): 1153–68. April 2014. doi:10.1016/j.bbamem.2013.10.004. PMID 24161712. https://dx.doi.org/10.1016%2Fj.bbamem.2013.10.004
- "Molecular mechanisms of inhibition of influenza by surfactant protein D revealed by large-scale molecular dynamics simulation". Biochemistry 52 (47): 8527–38. November 2013. doi:10.1021/bi4010683. PMID 24224757. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3927399
- "Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo". Nature Medicine 20 (2): 143–51. February 2014. doi:10.1038/nm.3443. PMID 24412922. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3966466
- "Towards a universal influenza vaccine: different approaches for one goal" (in En). Virology Journal 15 (1): 17. January 2018. doi:10.1186/s12985-017-0918-y. PMID 29370862. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5785881
- "An antibody that prevents the hemagglutinin low pH fusogenic transition". Virology 294 (1): 70–4. March 2002. doi:10.1006/viro.2001.1320. PMID 11886266. https://dx.doi.org/10.1006%2Fviro.2001.1320
- "Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses". Nature Structural & Molecular Biology 16 (3): 265–73. March 2009. doi:10.1038/nsmb.1566. PMID 19234466. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2692245
- "A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins". Science 333 (6044): 850–6. August 2011. doi:10.1126/science.1205669. PMID 21798894. https://dx.doi.org/10.1126%2Fscience.1205669
- "Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells". PLOS One 3 (12): e3942. 2008. doi:10.1371/journal.pone.0003942. PMID 19079604. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2596486
- "Antibody recognition of a highly conserved influenza virus epitope". Science 324 (5924): 246–51. April 2009. doi:10.1126/science.1171491. PMID 19251591. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2758658
- MICU, ALEXANDRU (2015-08-25). "Universal flu vaccine: research moves closer" (in en-US). http://www.zmescience.com/ecology/world-problems/effective-flu-vaccines-2788324.
- "Investigational hemagglutinin-targeted influenza virus inhibitors". Expert Opinion on Investigational Drugs 26 (1): 63–73. January 2017. doi:10.1080/13543784.2017.1269170. PMID 27918208. https://dx.doi.org/10.1080%2F13543784.2017.1269170




