| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Minh Tam Tran Thi | + 3423 word(s) | 3423 | 2020-11-25 08:50:28 | | | |
| 2 | Bruce Ren | Meta information modification | 3423 | 2020-12-03 03:51:55 | | |
Video Upload Options
Pseudomonas aeruginosa is an opportunistic human pathogen causing devastating acute and chronic infections in individuals with compromised immune systems. Its highly notorious persistence in clinical settings is attributed to its ability to form antibiotic-resistant biofilms. Biofilm is an architecture built mostly by autogenic extracellular polymeric substances which function as a scaffold to encase the bacteria together on surfaces, and to protect them from environmental stresses, impedes phagocytosis and thereby conferring the capacity for colonization and long-term persistence.
1. Introduction
Pseudomonas aeruginosa is an ubiquitous Gram-negative bacterium that causes nosocomial infections, as well as fatal infections in immunocompromised individuals, such as patients with cancer, post-surgery, severe burns or infected by human immunodeficiency virus (HIV) [1][2][3]. In 2017, P. aeruginosa was recognised as one of the most life-threatening bacteria and listed as priority pathogen for Research and Development of new antibiotics by the World Health Organization [4]. Common antimicrobial agents like antibiotics frequently exhibit limited efficacy due to adaptability and high intrinsic antibiotic resistance of P. aeruginosa, thus increasing mortality [5]. Additionally, treatment of these infections is also hindered by the P. aeruginosa ability to form biofilms which protect them from surrounding environmental stresses, impedes phagocytosis and thereby confers capacity for colonization and long-term persistence [6]. Such ability is promoted by effective cell-to-cell communications within the microbial communities of P. aeruginosa known as quorum sensing. As a result, highly structured biofilms can be formed which is often identified in patients with chronic infections, such as chronic lung infection, chronic wound infection and chronic rhinosinusitis [7]. It has been estimated that biofilms have a substantial bearing on over 90% of chronic wound infections, resulting in poor wound healing. In the United States alone, approximately 6.5 million patients were affected by chronic wound infections, which resulted in high health-care burden and devastating economic consequences estimated at over US$25 billion annually [8]. It is important therefore to diagnose P. aeruginosa infections at an early stage before biofilm development which could enhance the susceptibility of P. aeruginosa towards antimicrobial treatments. However, the increasing incidence of acute and persisting infections worldwide also highlights the need to develop therapeutic strategies as an alternative to traditional antibiotics, expectedly to disarm and eradicate this Gram-negative bacterium.
This review highlights the P. aeruginosa biofilms starting from its composition, structure and development processes to the extraordinary capabilities of P. aeruginosa to invade host immune system and escape antibiotic treatments via biofilm-mediated resistance which is regulated mainly by quorum sensing. In the context of challenges facing P. aeruginosa devastating infections, recent diagnostics and therapeutic strategies will be discussed.
2. Pseudomonas aeruginosa Biofilm
In nature, most bacteria can attach to different surfaces and form biofilms [9]. The biofilm is a complex aggregate of bacteria encased in a self-generated matrix of extracellular polymeric substances (EPS) and is one of the key strategies for the survival of species during unexpected changes of living conditions such as temperature fluctuation and nutrient availability [10]. Bacteria within a biofilm can escape host immune responses and resist antimicrobial treatments up to 1000 times more than their planktonic counterparts [11]. P. aeruginosa is a well-known biofilm former, which makes it an excellent model to study biofilm formation [12][13]. A resilient biofilm is a critical weapon for P. aeruginosa to compete, survive and dominate in the cystic fibrosis lung polymicrobial environment [14]. P. aeruginosa also effectively colonizes a variety of surfaces including medical materials (urinary catheters, implants, contact lenses, etc.) [12], and food industry equipment (mixing tanks, vats and tubing) [15]. Therefore, a greater understanding of the composition and structure of the biofilm, and the molecular mechanisms underlying the antimicrobial tolerance of bacteria growing within a biofilm, are vital for the design of effective strategies to manage, prevent and more importantly to eradicate biofilm-associated infections.
2.1. Biofilm Composition
The biofilm is a complex aggregate of bacteria encased in a self-generated matrix of extracellular polymeric substances (EPS) and is one of the key strategies for the survival of species against unexpected changes of living conditions such as temperature and nutrient availability [16][17].
It has been shown that P. aeruginosa biofilm matrix primarily encompasses polysaccharides, extracellular DNA (eDNA), proteins and lipids [18]. The matrix, which is responsible for more than 90% of biofilm biomass, acts as a scaffold for adhesion to biotic and abiotic surfaces and shelter for encased bacteria in harsh environmental conditions (antibiotics and host immune responses). It also provides a repertoire of public goods including essential nutrients, enzymes and cytosolic proteins for the biofilm community. The matrix also facilitates cell-to-cell communication [18][19][20].
The three exopolysaccharides, i.e., Psl, Pel and alginate, are tremendously involved in surface attachment, formation and the stability of biofilm architecture [21]. The roles of individual exopolysaccharides are discussed below.
Psl is a neutral pentasaccharide typically comprising d-glucose, d-mannose and l-rhamnose moieties [22][23]. This exopolysaccharide is necessary for adhesion of sessile cells (cells attached to a surface) to surfaces and cell-to-cell interactions during biofilm initiation of both nonmucoid and mucoid strains [24][25]. Psl has the following characteristics: (i) Psl is beneficial for biofilm communities, but not for unattached populations; (ii) better growth of non-Psl producing cells was observed in mixed biofilm with Psl producing cells [26]; (iii) during biofilm growth, Psl positive populations dominate Psl negative populations; and (iv) Psl nonproducers are unable to exploit Psl producers [26]. In a mature biofilm, Psl is located in peripheries of the mushroom-like structure where it helps maintain structural stability [22]. Increased Psl expression is linked to induction of cell aggregates in a liquid culture which is a phenotype observed in CF patients’ sputum [27]. Psl functions as a signalling molecule to promote the production of c-di-GMP (bis-(3′-5′)-cyclic dimeric guanosine monophosphate) whose level, if elevated, results in thicker and more robust biofilms [28]. Additionally, Psl shields biofilm bacteria from antimicrobials [21] and neutrophil phagocytosis [29], making it an effective defence to achieve persistent infection.
Pel is a cationic polysaccharide polymer of partially deacetylated N-acetyl-d-glucosamine and N-acetyl-d-galactosamine. Like Psl, Pel is an essential matrix component of biofilm in nonmucoid strains and is involved in the initiation of surface attachment, as well as maintenance of biofilm integrity [30][31]. Pel is responsible for the pellicle biofilm which is formed at the air-liquid interface of a static broth culture [32]. The synthesis of Psl and Pel are strain-specific and can be switched in response to surrounding conditions [33]. Pel promotes the tolerance to aminoglycoside antibiotics for biofilm-embedded bacteria [34]. Furthermore, Pel containing biofilms has been demonstrated to be refractory to the antibiotic colistin and less susceptible to killing mediated by neutrophils derived from human HL-60 cell lines [35]. Unlike Psl, Pel is not public goods and not available for Pel negative cells in both biofilm and unattached populations [26].
Alginate is predominately produced in the biofilm of mucoid Pseudomonas strains due to a mutation in mucA22 allele. The mucoid phenotypes are found mostly in CF isolates, signifying the conversion from acute to chronic infection [36][37]. Alginate is a negatively charged acetylated polymer consisting of mannuronic acid and guluronic acid residues [38]. A wide range of important functions of alginate including biofilm maturation, protection from phagocytosis and opsonization, and decreased diffusion of antibiotics through the biofilm has been well-documented [39][40][41]. The ratios between mannuronic acid and guluronic acid influence the viscoelastic properties of biofilms which lead to impairment of cough clearance in the lung of CF patients infected with P. aeruginosa [42][43][44].
Cell lysis releases DNA into the environment, and this extracellular DNA (eDNA) is one of the crucial constituents of biofilms. Cell lysis can be caused by environmental stress such as the antimicrobial treatment via the endolytic activity of endolysin Lys which is encoded in the R- and F-pyocin gene cluster. This can occur in both early development of biofilms and the planktonic phase where rod-shaped bacteria rapidly turn into round cells resulting from structural damage of the cell wall and followed by lysis. The released eDNA, cytosolic proteins and particularly RNA are subsequently encapsulated into membrane vesicles (MVs) which are formed via membrane fragments originating from the lysed cells [45]. eDNA can also be localized on the surface and the stalk of the mushroom-like microcolonies [46][47]. eDNA is involved in various processes: (i) as a nutrient source for bacteria in the biofilm; (ii) supporting cellular organization and alignment via twitching motility; (iii) as a cation chelator that interacts with divalent cations (Mg2+ and Ca2+) on the outer membrane and subsequently activates the type VI secretion system which disseminates virulence factors within the host; (iv) the deposition of eDNA caused biofilm environment and infection sites to become acidic, limiting the penetration of antimicrobial agents; and (v) the presence of eDNA in P. aeruginosa biofilms can influence the inflammatory process activated by neutrophil [48][49][50][51].
It is noteworthy to mention the intracellular biopolymer, polyhydroxyalkanoate (PHA), that does not directly play a structural role in the biofilm matrix but is produced in cells within the biofilm. PHA, a carbon and energy storage polymer, has been implicated in stress tolerance as well as attachment to abiotic surfaces such as glass [52]. Within microaerophilic/anaerobic zones of the biofilm, PHA might serve as an electron sink to maintain energy-generating metabolic processes.
2.2. Biofilm Development
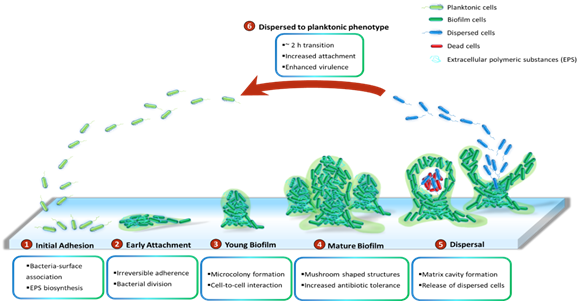
P.aeruginosa has been demonstrated to grow slowly as unattached cell aggregates under hypoxic and anoxic conditions, which are comparable to what has been observed in CF airways and chronic wounds [53]. Slow growth rates in the limited presence of oxygen are ascribed to antibiotic recalcitrance. Generally, biofilms of P. aeruginosa can be developed on abiotic surfaces, such as medical implants or industrial equipment. The biofilm development is divided into five distinct stages (Figure 1). Stage I: Bacterial cells adhere to a surface via support of cell appendages such as flagella and type IV pili [54][55]. The restricted flagellar movement has been implicated in mediating twitching motility and biosynthesis of exopolysaccharides required for surface association [56]. This adherence is reversible. A proteomic study with wild-type PAO1 concluded that the bacterial responses and biofilm formation are material specific. It is evident through records of the presence of specific bacterial proteins and their altered quantities when P. aeruginosa sense and react in response to a given surface [57]. Stage II: Bacterial cells undergo the switch from reversible to irreversible attachment. Stage III: Progressive propagation of attached bacteria into a more structured architecture, termed microcolonies. Stage IV: These microcolonies develop further into extensive three-dimensional mushroom-like structures, a hallmark of biofilm maturation. Stage V: In the center of the microcolony, matrix cavity is disrupted through cell autolysis for the liberation of dispersed cells followed by the transition from sessile to planktonic growth mode for seeding of uncolonized spaces (Stage VI), which allows the biofilm cycle to repeat [58]. It was recently demonstrated that endonuclease EndA is required for dispersion of existing biofilm via eDNA degradation [59]. The structure of the formed biofilms is influenced by the swarming motility with flat biofilms resulting from highly motile bacteria, while mushroom-shaped biofilms are generated by cells with low motility, and that the motility rate was nutrient specific [60].
Figure 1. Cycle of P. aeruginosa biofilm development. The development cycle is divided into six stages. Initially, the bacteria associate with the surface and produce extracellular polymeric substances (EPS) including proteins, polysaccharides, lipids and eDNA. Next, cell division and the transition of reversible attachment into irreversible take place. The following steps are the formation of microcolonies and the further development of these microcolonies into mushroom-shaped structures. Cell-to-cell interaction and production of virulence factors play essential roles in maturation and robustness of biofilms. Matrix cavity is then formed in the centre of microcolony via cell autolysis to disrupt the matrix for the liberation of the dispersed population. Finally, the released cells undergo an approximately 2 h transition into planktonic phenotypes which subsequently occupy uncolonized spaces.
While it is well-perceived that biofilm cells are physiologically different from their planktonic counterparts and more recalcitrant to antimicrobial treatments , little is known about intermediate events between attached forms and the free-swimming lifestyle which generates highly virulent detached cells. It is suggested that the transition from detachment to planktonic growth involves a distinct stage of biofilm development (Stage VI) (Figure 1). These cells, in contrast to both planktonic and sessile cells, possess distinct physiology and represent the conversion from chronic to acute infections. They turn into a planktonic phenotype after remaining in a 2-hour lag phase with decreased levels of pyoverdine and intracellular c-di-GMP [61][62]. Upregulation of virulence encoding genes and downregulation of iron uptake genes were observed in the dispersed population [63][64][65]. Both in vitro and in vivo experiments revealed that liberated cells are highly cytotoxic to macrophages, more sensitive to iron depletion and substantially virulent to nematode hosts relative to planktonic bacteria [63]. Notably, dispersed bacteria originating from biofilms treated with glycoside hydrolase rapidly disseminated and induced fatal septicaemia in a mouse chronic wound infection model [66].
2.3. Multispecies Biofilm
Generally, infections are not caused by monospecies alone but rather colonization of a complex polymicrobial community [67][68]. P. aeruginosa is often recognized as a co-colonizer along with other microbes such as Staphylococcus aureus (S. aureus), Burkholderia cenocepacia (B. cenocepacia) and Streptococcus parasanguinis (S. parasanguinis). For example, colonization of the biofilm-forming bacteria P. aeruginosa and S. aureus coinfects the lungs of CF patients and in diabetic and chronic wounds [69][70]. During co-infection, P. aeruginosa could sequester iron and nutrients through lysis of Gram-positive bacteria, including S. aureus, Streptococcus pneumoniae and Bacillus anthracis [67], as well as other Gram-negative bacteria Burkholderia cepacia [71].
In dual-species colonization containing P. aeruginosa and S. aureus, the presence of S. aureus derived peptidoglycan, N-acetylglucosamine (GlcNAc), induced P. aeruginosa to produce pyocyanin which functions as antimicrobials and toxins that could reduce the viability of S. aureus within the biofilm [68]. The transition from nonmucoid P. aeruginosa, which generate antimicrobial siderophores, rhamnolipids and 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), to mucoid phenotypes, which overproduce alginate, resulted in the decline of these exoproducts, leading to the cohabitant of P. aeruginosa and S. aureus [72]. S. aureus could also secrete extracellular adhesin known as staphylococcal protein A (SpA) which bound to Psl and type IV pilli on the P. aeruginosa cell surface, resulted in the inhibition of both biofilm formation of P. aeruginosa and phagocytotic activity of neutrophil towards P. aeruginosa [73]. P. aeruginosa has been demonstrated to outcompete S. aureus in the dual-species community by producing diguanylate cyclase, SiaD, which is activated by Psl, during the early stage of biofilm formation [74].
Similarly, synergistic interactions between P. aeruginosa and B. cenocepacia have been observed [75]. In planktonic co-cultures, P. aeruginosa predominated due to the production of secondary metabolites which inhibited the growth of B. cenocepacia. Moreover, co-existence in B. cenocepacia biofilm promoted biofilm biomass of P. aeruginosa. Co-infection of the two species was found to advance lung damage in a mouse model [75].
Although P. aeruginosa remains dominant in mixed-species biofilms by producing antimicrobial compounds which modulate the growth of other microorganisms, its pathogenesis was shown to be inhibited by oral streptococci strains during co-infection, resulting in improved CF lung conditions [76]. The oral commensal streptococci outcompeted P. aeruginosa by the production of hydrogen peroxide in the presence of nitrite [77]. Furthermore, it was shown that oral commensal S. parasanguinis could effectively exploit the exopolysaccharide alginate produced by P. aeruginosa CF isolate FRD1 strain to promote its biofilm in vitro through the mediation of the streptococcal surface adhesin BapA1. However, either adhesin BapA1 or Fap1 was adequate to reduce the colonization of alginate producing P. aeruginosa in Drosophila melanogaster [78].
3. Quorum sensing in biofilm development
The development of P. aeruginosa biofilms requires population-wide coordination of individual cells within bacterial communities [79]. P. aeruginosa uses multiple interconnected signal transduction pathways known as quorum sensing (QS), enabling the bacteria to communicate between the individual cells and ultimately orchestrate collective behaviour which is essential for the adaptation and survival of whole communities. P. aeruginosa enters into the QS mode in response to changes in cell density and environmental cues or stresses [80]. QS involves the production, secretion and accumulation of signalling molecules called autoinducers (AI) whose specificity and concentration are sensed by transcriptional regulators [81], resulting in the expressions of specific sets of genes on a population-wide scale. In addition to biofilm development, QS has been linked to the regulation of other physiological processes, including virulence-factor production, stress tolerance, metabolic adjustment and host-microbe interactions. Thus, understanding and controlling these chemical communication systems could lead to new targets for alternative or complementary treatments to conventional antimicrobials and antibiotics.
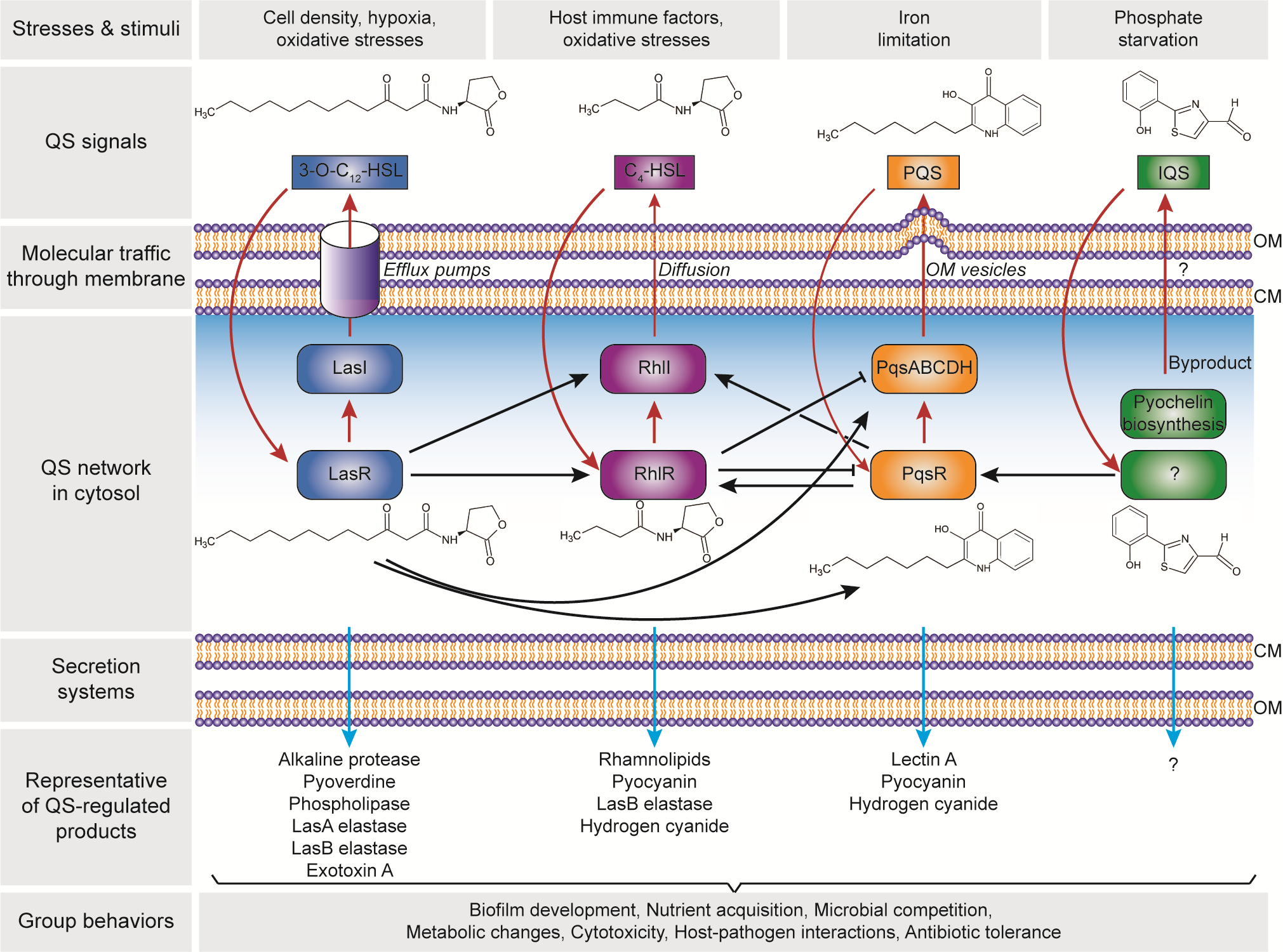
There are four distinct pathways in the QS circuits of P. aeruginosa, namely Las, Rhl, PQS and IQS that intracellularly produces their cognate AI molecules, i.e., N-3-oxo-dodecanoyl-ʟ-homoserine lactone (3O-C12-HSL), N-butyryl-ʟ-homoserine lactone (C4-HSL), 2-heptyl-3-hydroxy-4-quinolone (PQS) and 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (IQS), respectively (Figure 2). These QS circuits are organized in a hierarchy with the Las system at the top of the cascade [82]. Both Las and Rhl systems are triggered by an increased cell density at the preliminary exponential growth phase, whereas PQS and IQS systems are activated at late exponential growth phase [83] especially under iron limitation [84] and phosphate starvation conditions [85], respectively. The synthesized AIs undergo membrane trafficking directed to outside then inside of the cells, presumably mediated by free diffusion, efflux pumps or outer membrane vesicles [86]. The trafficked 3O-C12-HSL is then bound to the regulator protein LasR, and the formed complex activates lasI synthase gene, leading to the autoinduction feed-forward loop [87]. The LasR–3O-C12-HSL also induces the expression of rhlR and rhlI genes as well as the pqsR and pqsABCDH genes which encode the Rhl [82] and the PQS [88] systems, respectively. Similar to the Las system, the RhlR–C4-HSL complex induces rhlI gene expression that activates the second autoinduction feed-forward loop [89]. In the PQS system, the PqsR–PQS complex activates pqsABCDH genes as well as feeds back to induce rhlRI gene expression [90]. The expression of both pqsR and pqsABCDH genes can be inhibited by RhlR, which has been suggested as a way to control the correct ratio between 3-oxo-C12-HSL and C4-HSL, hence controlling the activation of PQS pathway [91]. The identification of IQS system was relatively new as compared to the other QS systems. In the IQS system, the identity of the transcriptional regulator is still unknown although its binding to the IQS has been found to activate the pqsR gene [92][93]. In addition, the IQS molecule was proposed to be enzymatically produced from the proteins encoded by ambBCDE genes [92]. However, the recent identification of the IQS molecule (an aeruginaldehyde) revealed that it is a byproduct of the siderophore pyochelin biosynthesis [94][95]. On the other hand, ambBCDE genes encode for proteins involved in the biosynthesis of the anti-metabolite L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) [96].
Figure 2. Hierarchical quorum-sensing (QS) network in Pseudomonas aeruginosa. The four QS pathways are activated in response to the cell density and environmental stimuli, with four autoinducer synthases including LasI, RhlI, PqsABCDH and AmbBCDE that produce N-3-oxo-dodecanoyl-ʟ-homoserine lactone (3O-C12-HSL), N-butyryl-ʟ-homoserine lactone (C4-HSL), 2-heptyl-3-hydroxy-4-quinolone (PQS) and 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (IQS), respectively. Note: the autoinduction is depicted in red arrows; the receptor for IQS is still unknown. The QS products are secreted through the cell membrane, that control the group behaviours and essential for the adaptation, survival and pathogenicity of P. aeruginosa. Abbreviation: CM, cytoplasmic membrane; OM, outer membrane.
The Las, Rhl and PQS systems in the QS network of P. aeruginosa play important roles in the production of the functional elements that have an impact on biofilm development (Figure 2). These include rhamnolipid [97][98], pyoverdine [99], pyocyanin [100][101], Pel polysaccharides[102], and lectins [103][104]. Rhamnolipid is a rhamnose-containing glycolipidic compound (i.e., biosurfactant) that functions to preserve the pores and channels between microcolonies, enabling the passage of liquid and nutrients within mature biofilms. Pyoverdine can sequester iron in the environment and delivery it to the cell which is an essential component for biofilm development. In an environment where iron is limited, twitching motility is more favoured than sessile growth and thus inhibiting biofilm formation [105]. Pyocyanin is a secondary metabolite with a cytotoxicity effect, thereby inducing cell lysis and releasing the cells’ DNA to extracellular space (i.e., eDNA – one of the biofilm components). Pyocyanin can bind to the eDNA and causes an increase in solution viscosity, thus also increasing the physicochemical interactions between biofilm matrices and the surrounding environment as well as promoting cellular aggregation. Pel polysaccharides can also interact with eDNA through cationic-anionic interactions within the biofilm matrix, strengthening the biofilm structure. Lectins are soluble proteins located in the outer membrane which consist of two forms, i.e., LecA (that binds to galactose and its derivatives) and LecB (that binds to fucose, mannose and mannose-containing oligosaccharides). Such adhesive properties of lectins facilitate the retention of both cells and exopolysaccharides in a growing biofilm, contributing to the biofilm structure, as well as adhesion to biological surfaces such as epithelium and mucosa. Collectively, such molecular and cellular interactions in combination with other polymeric substances lead to the establishment of a robust and mature biofilm.
References
- Gale, M.J.; Maritato, M.S.; Chen, Y.-L.; Abdulateef, S.S.; Ruiz, J.E. Pseudomonas aeruginosa causing inflammatory mass of the nasopharynx in an immunocompromised HIV infected patient: A mimic of malignancy. IDCases 2015, 2, 40–43, doi:10.1016/j.idcr.2015.01.004.
- Wu, W.; Jin, Y.; Bai, F.; Jin, S. Chapter 41—Pseudomonas Aeruginosa. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 753–767. doi:10.1016/B978-0-12-397169-2.00041-X.
- Gomila, A.; Carratalà, J.; Badia, J.M.; Camprubí, D.; Piriz, M.; Shaw, E.; Diaz-Brito, V.; Espejo, E.; Nicolás, C.; Brugués, M.; et al. Preoperative oral antibiotic prophylaxis reduces Pseudomonas aeruginosa surgical site infections after elective colorectal surgery: A multicenter prospective cohort study. BMC Infect. Dis. 2018, 18, 507–507, doi:10.1186/s12879-018-3413-1.
- World Health Organization. Prioritization of pathogens to guide discovery,research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis; World Health Organization: Geneva, Switzerland, 2017.
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192, doi:10.1016/j.biotechadv.2018.11.013.
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39, doi:10.3389/fcimb.2017.00039.
- Romling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561, doi:10.1111/joim.12004.
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen 2009, 17, 763–771, doi:10.1111/j.1524-475X.2009.00543.x.
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890, doi:10.3201/eid0809.020063.
- Rollet, C.; Gal, L.; Guzzo, J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: A comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 290, 135–142, doi:10.1111/j.1574-6968.2008.01415.x.
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007, doi:10.1128/AAC.45.4.999-1007.2001.
- Ghafoor, A.; Hay, I.D.; Rehm, B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246, doi:10.1128/AEM.00637-11.
- Crespo, A.; Blanco-Cabra, N.; Torrents, E. Aerobic Vitamin B12 Biosynthesis Is Essential for Pseudomonas aeruginosa Class II Ribonucleotide Reductase Activity During Planktonic and Biofilm Growth. Front. Microbiol. 2018, 9, 986, doi:10.3389/fmicb.2018.00986.
- Oluyombo, O.; Penfold, C.N.; Diggle, S.P. Competition in Biofilms between Cystic Fibrosis Isolates of Pseudomonas aeruginosa Is Shaped by R-Pyocins. mBio 2019, 10, e01828-18, doi:10.1128/mBio.01828-18.
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordonez, A. New Weapons to Fight Old Enemies: Novel Strategies for the (Bio)control of Bacterial Biofilms in the Food Industry. Front. Microbiol. 2016, 7, 1641, doi:10.3389/fmicb.2016.01641.
- Moradali, M.F.; Rehm, B.H.A. Bacterial biopolymers: From pathogenesis to advanced materials. Nat. Rev. Microbiol. 2020, 18, 195–210, doi:10.1038/s41579-019-0313-3.
- Rehm, B.H.A. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592, doi:10.1038/nrmicro2354.
- Strempel, N.; Neidig, A.; Nusser, M.; Geffers, R.; Vieillard, J.; Lesouhaitier, O.; Brenner-Weiss, G.; Overhage, J. Human host defense peptide LL-37 stimulates virulence factor production and adaptive resistance in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e82240, doi:10.1371/journal.pone.0082240.
- Jackson, K.D.; Starkey, M.; Kremer, S.; Parsek, M.R.; Wozniak, D.J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 2004, 186, 4466–4475, doi:10.1128/JB.186.14.4466-4475.2004.
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648, doi:10.1016/j.mib.2007.09.010.
- Billings, N.; Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 2013, 9, e1003526, doi:10.1371/journal.ppat.1003526.
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009, 5, e1000354, doi:10.1371/journal.ppat.1000354.
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638, doi:10.1111/j.1365-2958.2009.06795.x.
- Ma, L.; Wang, S.; Wang, D.; Parsek, M.R.; Wozniak, D.J. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 2012, 65, 377–380, doi:10.1111/j.1574-695X.2012.00934.x.
- Jones, C.J.; Wozniak, D.J. Psl Produced by Mucoid Pseudomonas aeruginosa Contributes to the Establishment of Biofilms and Immune Evasion. mBio 2017, 8, e00864-17, doi:10.1128/mBio.00864-17.
- Irie, Y.; Roberts, A.E.L.; Kragh, K.N.; Gordon, V.D.; Hutchison, J.; Allen, R.J.; Melaugh, G.; Bjarnsholt, T.; West, S.A.; Diggle, S.P. The Pseudomonas aeruginosa PSL Polysaccharide Is a Social but Noncheatable Trait in Biofilms. mBio 2017, 8, e00374-17, doi:10.1128/mBio.00374-17.
- Staudinger, B.J.; Muller, J.F.; Halldórsson, S.; Boles, B.; Angermeyer, A.; Nguyen, D.; Rosen, H.; Baldursson, O.; Gottfreðsson, M.; Guðmundsson, G.H.; et al. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 2014, 189, 812–824, doi:10.1164/rccm.201312-2142OC.
- Irie, Y.; Borlee, B.R.; O'Connor, J.R.; Hill, P.J.; Harwood, C.S.; Wozniak, D.J.; Parsek, M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2012, 109, 20632–20636, doi:10.1073/pnas.1217993109.
- Mishra, M.; Byrd, M.S.; Sergeant, S.; Azad, A.K.; Parsek, M.R.; McPhail, L.; Schlesinger, L.S.; Wozniak, D.J. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012, 14, 95–106, doi:10.1111/j.1462-5822.2011.01704.x.
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339, doi:10.1128/JB.02150-12.
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358, doi:10.1073/pnas.1503058112.
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690.
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2011, 14, 1913–1928, doi:10.1111/j.1462-2920.2011.02657.x.
- Yang, L.; Hu, Y.; Liu, Y.; Zhang, J.; Ulstrup, J.; Molin, S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 2011, 13, 1705–1717, doi:10.1111/j.1462-2920.2011.02503.x.
- Baker, P.; Hill, P.J.; Snarr, B.D.; Alnabelseya, N.; Pestrak, M.J.; Lee, M.J.; Jennings, L.K.; Tam, J.; Melnyk, R.A.; Parsek, M.R.; et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016, 2, e1501632, doi:10.1126/sciadv.1501632.
- Ciofu, O.; Tolker-Nielsen, T.; Jensen, P. Østrup; Wang, H.; Høiby, N. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv. Drug Deliv. Rev. 2015, 85, 7–23, doi:10.1016/j.addr.2014.11.017.
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Genet. 2012, 10, 841–851, doi:10.1038/nrmicro2907.
- Evans, L.R.; Linker, A. Production and Characterization of the Slime Polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924, doi:10.1128/jb.116.2.915-924.1973.
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protectsPseudomonas aeruginosabiofilms by limiting the penetration of tobramycin. Environ. Microbiol. 2013, 15, 2865–2878, doi:10.1111/1462-2920.12155.
- Hay, I.D.; Rehman, Z.U.; Moradali, M.F.; Wang, Y.; Rehm, B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650, doi:10.1111/1751-7915.12076.
- Hay, I.D.; Rehman, Z.U.; Ghafoor, A.; Rehm, B.H.A. Bacterial biosynthesis of alginates. J. Chem. Technol. Biotechnol. 2010, 85, 752–759, doi:10.1002/jctb.2372.
- Gloag, E.S.; German, G.K.; Stoodley, P.; Wozniak, D.J. Viscoelastic properties of Pseudomonas aeruginosa variant biofilms. Sci. Rep. 2018, 8, 1–11, doi:10.1038/s41598-018-28009-5.
- Wloka, M.; Rehage, H.; Flemming, H.-C.; Wingender, J. Structure and rheological behaviour of the extracellular polymeric substance network of mucoid Pseudomonas aeruginosa biofilms. Biofilms 2005, 2, 275–283, doi:10.1017/s1479050506002031.
- Rehm, B.H.A.; Valla, S. Bacterial alginates: biosynthesis and applications. Appl. Microbiol. Biotechnol. 1997, 48, 281–288, doi:10.1007/s002530051051.
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 11220, doi:10.1038/ncomms11220.
- Chiang, W.-C.; Nilsson, M.; Jensen, P. Østrup; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA Shields against Aminoglycosides in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361, doi:10.1128/aac.00001-13.
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.C.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128, doi:10.1111/j.1365-2958.2005.05008.x.
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 60, 544–553, doi:10.1128/aac.01650-15.
- Wilton, M.; Wong, M.J.Q.; Tang, L.; Liang, X.; Moore, R.; Parkins, M.D.; Lewenza, S.; Dong, T.G. Chelation of Membrane-Bound Cations by Extracellular DNA Activates the Type VI Secretion System in Pseudomonas aeruginosa. Infect. Immun. 2016, 84, 2355–2361, doi:10.1128/iai.00233-16.
- Gloag, E.S.; Turnbull, L.; Huang, A.; Vallotton, P.; Wang, H.; Nolan, L.M.; Mililli, L.; Hunt, C.; Lu, J.; Osvath, S.R.; et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. 2013, 110, 11541–11546.
- Bass, J.I.F.; Russo, D.M.; Gabelloni, M.L.; Geffner, J.R.; Giordano, M.; Catalano, M.; Zorreguieta, Á.; Trevani, A.S. Extracellular DNA: A Major Proinflammatory Component of Pseudomonas aeruginosaBiofilms. J. Immunol. 2010, 184, 6386–6395, doi:10.4049/jimmunol.0901640.
- Pham, T.H.; Webb, J.S.; Rehm, B.H.A. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology 2004, 150, 3405–3413, doi:10.1099/mic.0.27357-0.
- Sønderholm, M.; Kragh, K.N.; Koren, K.; Jakobsen, T.H.; Darch, S.E.; Alhede, M.; Jensen, P. Østrup; Whiteley, M.; Kühl, M.; Bjarnsholt, T. Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics. Appl. Environ. Microbiol. 2017, 83, doi:10.1128/aem.00113-17.
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosabiofilm development. Mol. Microbiol. 1998, 30, 295–304, doi:10.1046/j.1365-2958.1998.01062.x.
- Klausen, M.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68, doi:10.1046/j.1365-2958.2003.03677.x.
- Hickman, J.W.; Harwood, C.S. Identification of FleQ fromPseudomonas aeruginosaas a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008, 69, 376–389, doi:10.1111/j.1365-2958.2008.06281.x.
- Guilbaud, M.; Bruzaud, J.; Bouffartigues, E.; Orange, N.; Guillot, A.; Aubert-Frambourg, A.; Monnet, V.; Herry, J.-M.; Chevalier, S.; Bellon-Fontaine, M.-N. Proteomic Response of Pseudomonas aeruginosa PAO1 Adhering to Solid Surfaces. Front. Microbiol. 2017, 8, 1465, doi:10.3389/fmicb.2017.01465.
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 1–17, doi:10.1155/2015/759348.
- Cherny, K.E.; Sauer, K. Pseudomonas aeruginosa Requires the DNA-Specific Endonuclease EndA To Degrade Extracellular Genomic DNA To Disperse from the Biofilm. J. Bacteriol. 2019, 201, doi:10.1128/jb.00059-19.
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277, doi:10.1111/j.1365-2958.2006.05421.x.
- Chua, S.L.; Tan, S.Y.-Y.; Rybtke, M.T.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Tolker-Nielsen, T.; Yang, L.; Givskov, M. Bis-(3′-5′)-Cyclic Dimeric GMP Regulates Antimicrobial Peptide Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 2066–2075, doi:10.1128/aac.02499-12.
- Chua, S.L.; Hultqvist, L.D.; Yuan, M.; Rybtke, M.; E Nielsen, T.; Givskov, M.; Tolker-Nielsen, T.; Yang, L. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm–dispersed cells via c-di-GMP manipulation. Nat. Protoc. 2015, 10, 1165–1180, doi:10.1038/nprot.2015.067.
- Chua, S.L.; Liu, Y.; Yam, J.K.H.; Chen, Y.; Vejborg, R.M.; Tan, B.G.C.; Kjelleberg, S.; Tolker-Nielsen, T.; Givskov, M.; Yang, L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014, 5, 4462, doi:10.1038/ncomms5462.
- Chambers, J.R.; Cherny, K.E.; Sauer, K. Susceptibility of Pseudomonas aeruginosa Dispersed Cells to Antimicrobial Agents Is Dependent on the Dispersion Cue and Class of the Antimicrobial Agent Used. Antimicrob. Agents Chemother. 2017, 61, e00846-17, doi:10.1128/aac.00846-17.
- Li, Y.; Petrova, O.E.; Su, S.; Lau, G.W.; Panmanee, W.; Na, R.; Hassett, D.J.; Davies, D.G.; Sauer, K. BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1004168, doi:10.1371/journal.ppat.1004168.
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 1–7, doi:10.1038/s41598-018-29121-2.
- Mashburn, L.M.; Jett, A.M.; Akins, D.R.; Whiteley, M. Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J. Bacteriol. 2005, 187, 554–566, doi:10.1128/jb.187.2.554-566.2005.
- Korgaonkar, A.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. 2012, 110, 1059–1064, doi:10.1073/pnas.1214550110.
- Willner, D.L.; Haynes, M.R.; Furlan, M.; Schmieder, R.; Lim, Y.W.; Rainey, P.B.; Rohwer, F.; Conrad, D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012, 6, 471–474, doi:10.1038/ismej.2011.104.
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108, doi:10.1038/nrmicro821.
- Weaver, V.B.; Kolter, R. Burkholderia spp. Alter Pseudomonas aeruginosa Physiology through Iron Sequestration. J. Bacteriol. 2004, 186, 2376–2384, doi:10.1128/jb.186.8.2376-2384.2004.
- Limoli, D.H.; Whitfield, G.B.; Kitao, T.; Ivey, M.L.; Davis, M.R., Jr.; Grahl, N.; Hogan, D.A.; Rahme, L.G.; Howell, P.L.; O’Toole, G.A.; et al. Pseudomonas aeruginosaAlginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. mBio 2017, 8, e00186-17, doi:10.1128/mbio.00186-17.
- Armbruster, C.R.; Wolter, D.J.; Mishra, M.; Hayden, H.S.; Radey, M.C.; Merrihew, G.; MacCoss, M.J.; Burns, J.; Wozniak, D.J.; Parsek, M.R.; et al. Staphylococcus aureus Protein A Mediates Interspecies Interactions at the Cell Surface of Pseudomonas aeruginosa. mBio 2016, 7, e00538-16, doi:10.1128/mbio.00538-16.
- Chew, S.C.; Yam, J.K.H.; Matysik, A.; Seng, Z.J.; Klebensberger, J.; Givskov, M.; Doyle, P.; Rice, S.A.; Yang, L.; Kjelleberg, S. Matrix Polysaccharides and SiaD Diguanylate Cyclase Alter Community Structure and Competitiveness of Pseudomonas aeruginosa during Dual-Species Biofilm Development with Staphylococcus aureus. mBio 2018, 9, doi:10.1128/mbio.00585-18.
- Bragonzi, A.; Farulla, I.; Paroni, M.; Twomey, K.B.; Pirone, L.; Lorè, N.I.; Bianconi, I.; Dalmastri, C.; Ryan, R.P.; Bevivino, A. Modelling Co-Infection of the Cystic Fibrosis Lung by Pseudomonas aeruginosa and Burkholderia cenocepacia Reveals Influences on Biofilm Formation and Host Response. PLoS ONE 2012, 7, e52330, doi:10.1371/journal.pone.0052330.
- Filkins, L.M.; Hampton, T.H.; Gifford, A.H.; Gross, M.J.; Hogan, D.A.; Sogin, M.L.; Morrison, H.G.; Paster, B.J.; O’Toole, G.A. Prevalence of Streptococci and Increased Polymicrobial Diversity Associated with Cystic Fibrosis Patient Stability. J. Bacteriol. 2012, 194, 4709–4717, doi:10.1128/jb.00566-12.
- Scoffield, J.A.; Wu, H. Oral Streptococci and Nitrite-Mediated Interference of Pseudomonas aeruginosa. Infect. Immun. 2015, 83, 101–107, doi:10.1128/iai.02396-14.
- Scoffield, J.A.; Duan, D.; Zhu, F.; Wu, H. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLoS Pathog. 2017, 13, e1006300, doi:10.1371/journal.ppat.1006300.
- Yan, S.; Wu, G. Can Biofilm Be Reversed Through Quorum Sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 1582, doi:10.3389/fmicb.2019.01582.
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Genet. 2019, 17, 371–382, doi:10.1038/s41579-019-0186-5.
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275, doi:10.1128/jb.176.2.269-275.1994.
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41, doi:10.1007/s13238-014-0100-x.
- Choi, Y.; Park, H.-Y.; Park, S.J.; Park, S.-J.; Kim, S.-K.; Ha, C.; Im, S.-J.; Lee, J.-H. Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells 2011, 32, 57–65, doi:10.1007/s10059-011-2322-6.
- Oglesby, A.G.; Farrow, J.M.I.; Lee, J.-H.; Tomaras, A.P.; Greenberg, E.P.; Pesci, E.C.; Vasil, M.L. The Influence of Iron on Pseudomonas aeruginosa Physiology: A regulatory link between iron and quorum sensing. J. Biol. Chem. 2008, 283, 15558–15567, doi:10.1074/jbc.m707840200.
- Rampioni, G.; Falcone, M.; Heeb, S.; Frangipani, E.; Fletcher, M.P.; Dubern, J.-F.; Visca, P.; Leoni, L.; Cámara, M.; Williams, P. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1006029, doi:10.1371/journal.ppat.1006029.
- Mashburn, L.M.; Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nat. Cell Biol. 2005, 437, 422–425, doi:10.1038/nature03925.
- Seed, P.C.; Passador, L.; Iglewski, B.H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: An autoinduction regulatory hierarchy. J. Bacteriol. 1995, 177, 654–659, doi:10.1128/jb.177.3.654-659.1995.
- Déziel, E.; Lépine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. 2004, 101, 1339–1344, doi:10.1073/pnas.0307694100.
- Winson, M.K.; Camara, M.; Latifi, A.; Foglino, M.; Chhabra, S.R.; Daykin, M.; Bally, M.; Chapon, V.; Salmond, G.P.; Bycroft, B.W. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 1995, 92, 9427–9431, doi:10.1073/pnas.92.20.9427.
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas Quinolone Signal Regulates rhl Quorum Sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708, doi:10.1128/jb.182.10.2702-2708.2000.
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. 2001, 98, 14613–14618, doi:10.1073/pnas.251465298.
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343, doi:10.1038/nchembio.1225.
- Meng, X.; Ahator, S.D.; Zhang, L.-H. Molecular Mechanisms of Phosphate Stress Activation of Pseudomonas aeruginosa Quorum Sensing Systems. mSphere 2020, 5, 119–120, doi:10.1128/msphere.00119-20.
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Christophersen, C.; Hammerich, O. Structure Revision of N-Mercapto-4-formylcarbostyril Produced by Pseudomonas fluorescens G308 to 2-(2-Hydroxyphenyl)thiazole-4-carbaldehyde [aeruginaldehyde]. Nat. Prod. Commun. 2014, 9, 789–794, doi:10.1177/1934578x1400900615.
- Trottmann, F.; Franke, J.; Ishida, K.; Garcia-Altares, M.; Hertweck, C. A Pair of Bacterial Siderophores Releases and Traps an Intercellular Signal Molecule: An Unusual Case of Natural Nitrone Bioconjugation. Angew. Chem. Int. Ed. 2018, 58, 200–204, doi:10.1002/anie.201811131.
- Murcia, N.E.; Elee, X.; Ewaridel, P.; Emaspoli, A.; Imker, H.J.; Echai, T.; Walsh, C.T.; Reimmann, C. The Pseudomonas aeruginosa antimetabolite L -2-amino-4-methoxy-trans-3-butenoic acid (AMB) is made from glutamate and two alanine residues via a thiotemplate-linked tripeptide precursor. Front. Microbiol. 2015, 6, 170, doi:10.3389/fmicb.2015.00170.
- Pamp, S.J.; Tolker-Nielsen, T. Multiple Roles of Biosurfactants in Structural Biofilm Development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539, doi:10.1128/jb.01515-06.
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767, doi:10.1128/jb.179.18.5756-5767.1997.
- Banin, E.; Vasil, M.L.; Greenberg, E.P. From The Cover: Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. 2005, 102, 11076–11081, doi:10.1073/pnas.0504266102.
- Das, T.; Kutty, S.K.; Kumar, N.; Manefield, M. Pyocyanin Facilitates Extracellular DNA Binding to Pseudomonas aeruginosa Influencing Cell Surface Properties and Aggregation. PLoS ONE 2013, 8, e58299, doi:10.1371/journal.pone.0058299.
- Das, T.; Kutty, S.K.; Tavallaie, R.; Ibugo, A.I.; Panchompoo, J.; Sehar, S.; Aldous, L.; Yeung, A.W.S.; Thomas, S.R.; Kumar, N.; et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci. Rep. 2015, 5, srep08398, doi:10.1038/srep08398.
- Sakuragi, Y.; Kolter, R. Quorum-Sensing Regulation of the Biofilm Matrix Genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386, doi:10.1128/jb.00137-07.
- Da Silva, D.P.; Matwichuk, M.L.; Townsend, D.O.; Reichhardt, C.; Lamba, D.; Wozniak, D.J.; Parsek, M.R. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019, 10, 1–11, doi:10.1038/s41467-019-10201-4.
- Diggle, S.P.; Stacey, R.E.; Dodd, C.; Cámara, M.; Williams, P.; Winzer, K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 2006, 8, 1095–1104, doi:10.1111/j.1462-2920.2006.001001.x.
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30, doi:10.1016/j.tim.2006.11.004.