The glycation of various biomolecules is the root cause of many pathological conditions associated with diabetic nephropathy and end-stage kidney disease. Glycation imbalances metabolism and increases renal cell injury. Numerous therapeutic measures have narrowed down the adverse effects of endogenous glycation, but efficient and potent measures are miles away. Recent advances in the identification and characterization of noncoding RNAs, especially the long noncoding RNAs (lncRNAs), have opened a mammon of new biology to explore the mitigations for glycation-associated diabetic nephropathy. Furthermore, tissue-specific distribution and condition-specific expression make lncRNA a promising key for second-generation therapeutic interventions.
1. Introduction
With the advancement in high-throughput sequencing technologies, including RNA sequencing and next-generation sequencing, researchers’ understanding of genome and transcriptome has improved remarkably. The advanced genome study started by several consortia such as the Human Genome Project, Encyclopedia of DNA element (ENCODE), and Functional Annotation of the mouse/mammalian genome (FANTOM) has eventually publicized that only a small portion (3%) of the complete genome relates to known protein-coding genes. The remaining amount (97%) is the so-called ‘unchartered territory of noncoding RNA’ with small fractions decoded
[1]. The established understanding of gene regulation in nature revolves around the central dogma of life (DNA → mRNA → protein); however, the rediscovery of noncoding RNA (ncRNA) genes and their role in the regulation of protein-coding genes has stirred the world
[2]. The advanced genomic studies pointed out that most RNA transcripts are noncoding and identified about 35,000 ncRNA transcripts exhibiting signatures such as mRNA, including polyadenylation, capping, and splicing
[3].
Moreover, due to their mRNA-like features, these newly identified ncRNA transcripts are difficult to distinguish from protein-coding transcripts. Despite these cumbersome challenges, various phases of these consortia identified almost around 27,919 long noncoding RNAs (lncRNAs)
[4]. These findings suggested that human genome transcription is ubiquitous and produces thousands of ncRNA transcripts
[5]. The ncRNAs can be further grouped into two classes based on their nucleotide number. One group consists of RNA genes with fewer than 200 nucleotides (microRNAs, small interfering RNAs, Piwi-interacting RNAs, and small nucleolar RNAs)
[6], while the other group comprises RNA genes with more than 200 nucleotides, called lncRNAs
[7]. The lncRNAs are present abundantly in both animals and plants, and their presence in the plant kingdom can be seen as an example of combinatorial transcriptional regulation
[8]. The expression patterns of lncRNAs depend on many biological factors, including high glucose (HG), free fatty acids, growth factors, and inflammatory cytokines; hence, they are considered biomarkers in many pathological conditions (
Table 1)
[9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24].
Table 1. LncRNAs involved in various diabetes-associated complications.
| Diabetic Complications |
lncRNA Involved |
Mode of Action |
References |
| Diabetic neuropathy |
lncRNA NEAT1 |
Regulate disease progression by targeting two miRNAs, miR-183-5p and miR-433-3p. |
[9] |
| lncRNA TP73-AS1 |
Sponges decreases miR-142 and upregulates HMGB1 expression as well as promotes cell proliferation. Its silencing decreases neuropathic pain. |
[10] |
| Diabetic retinopathy |
lncRNA HOTTIP |
Induces p38/MAPK signaling and promotes retinal cell inflammatory response and diabetic retinopathy progression. |
[11] |
| lncRNA BANCR |
Regulates cell apoptosis |
[12] |
| lncRNA MEG3 |
Regulates VEGF and TGF-β1 expressions |
[13] |
| Inflammatory diabetes complications |
lncRNA DRAIR |
Regulates macrophages/monocyte pro/anti-inflammatory phenotypes in T2DM. Down-regulated by diabetogenic factors |
[14] |
| Diabetic wound healing |
lncRNA URIDS |
Impairs collagen production and crosslinking by interacting with Plod1 and delays wound healing |
[15] |
| lncRNA MALAT1 |
Increases wound healing and upregulate fibroblast activation in diabetic mice by activating the HIF-1α signaling pathway. |
[16] |
| Diabetic cardiomyopathy |
lncRNA HOTAIR |
Downregulates DCM effects by activating SIRT1 expressions and sponging miR-34a. |
[17] |
| lncRNA Kcnq1ot1 |
Promotes pyroptosis by regulating expressions of miR-214-3p and caspase-1 |
[18] |
| lncRNA H19 |
Regulates high glucose-induced apoptosis by targeting VDAC1. It also improves left ventricular function when overexpressed. |
[19] |
| lncRNA Crende |
Negatively regulates cardiac fibroblast differentiation. Also, its expression is induced by Smad3 in cardiac fibroblasts. It inhibits myofibroblastic gene transcription. |
[20] |
| lncRNA TUG1 |
Its knockdown lessened DCM-induced hypertrophy and diastolic function. Also, its silencing upregulates the expression of some miRNAs |
[21] |
| Diabetic nephropathy |
lncRNA Blnc1 |
Attenuates renal fibrosis and inflammation and affects oxidative stress by NF-κB and NRF2/HO-1 pathways |
[22] |
| lncRNA TCF7 |
It acts as a sponge against miR-200c and triggers endoplasmic reticulum stress in patients with DN. |
[23] |
| lncRNA Gas5 |
Alleviates cell proliferation and fibrosis sponging miR-221 and upregulates SIRT1 |
[24] |
1.1. Structural Properties of LncRNAs
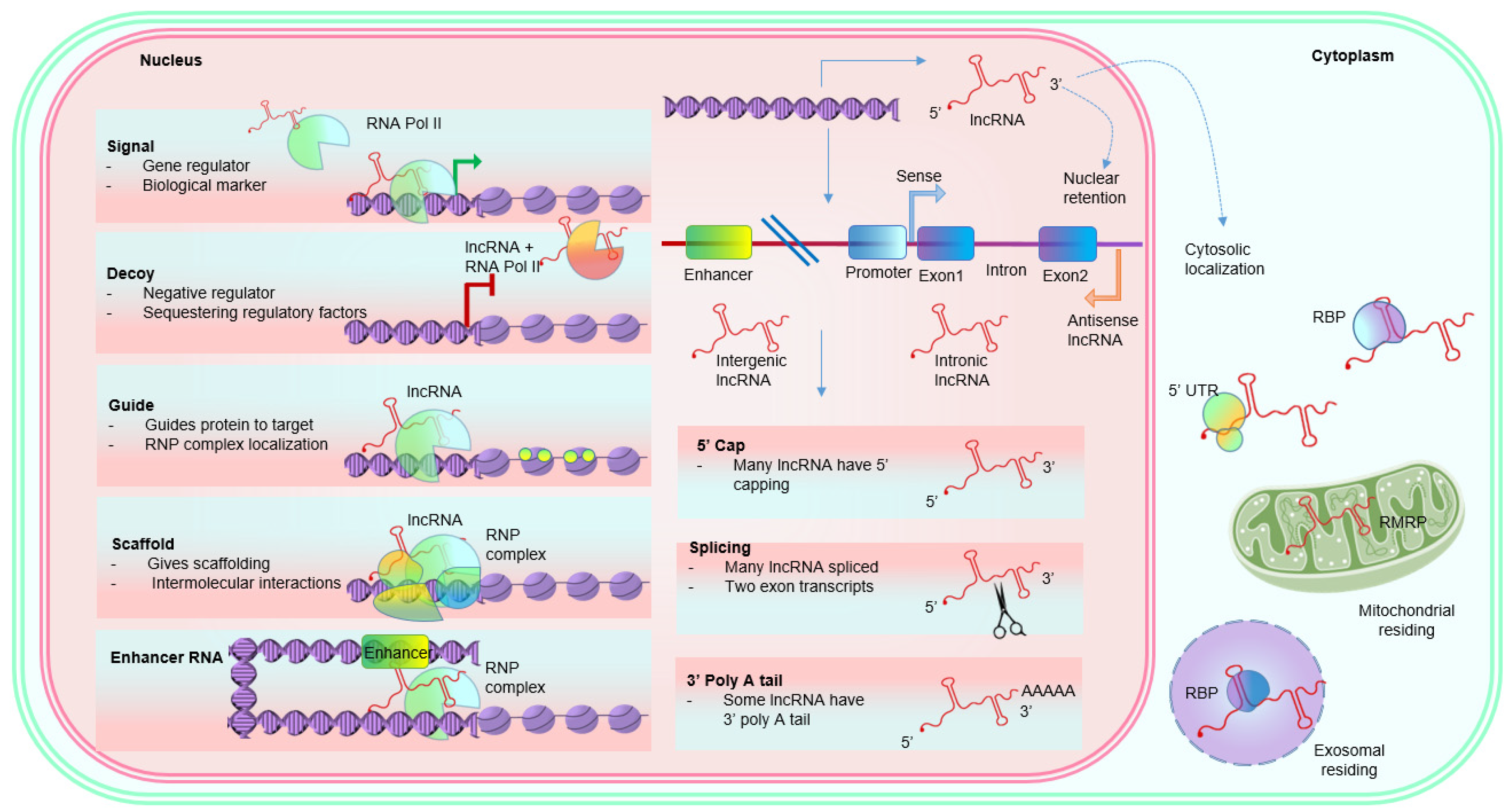
Many lncRNAs transcribe and process like mRNAs (
Figure 1). The generative pathways (histone-modification patterns, splicing, spliceosome signals, and exons/introns segments) for lncRNAs are similar to those of protein-coding genes
[25]. Nevertheless, unlike mRNAs, a significant portion of lncRNAs are predominantly located in the nucleus, then in the cytoplasm
[26]. Additionally, lncRNAs are broadly divided into categories based on genomic locations, such as 1. Intergenic lncRNAs (located between two separate genes), 2. Intronic lncRNAs (located between two exons), 3. Sense, or 4. Antisense lncRNAs (located on the same or opposite strands of protein-coding transcripts) and 5. Bidirectional lncRNAs
[27][28]. This location-based classification is helpful for structural recognitions and annotations; however, lncRNAs’ biological functions are equally essential for more accurate classification. The majority of the lncRNAs have a half-life of more than 16 h. The intergenic and
cis-antisense lncRNAs are more stable than intronic lncRNAs.
Figure 1. Archetypes and characteristics of lncRNAs along with their nuclear or cytoplasmic localizations. The various archetypes explain their mechanism of action in a non-mutually exclusive fashion. LncRNAs exhibit post-transcriptional signatures such as mRNA. Depending upon the splicing, lncRNA can either retain in the nucleus or localize in the cytosol. Abbreviations: LncRNAs, long noncoding RNAs; RNP, ribonucleoprotein; UTR, untranslated region; RMRP, RNA component of mitochondrial RNA-processing endoribonuclease; RBP, RNA-binding protein.
1.2. Functional Properties of LncRNAs
1.2.1. Role in Transcription Regulation
Gene regulation by lncRNAs can be facilitated through one of the two non-mutually exclusive ways: 1. By regulating adjacent gene expression and 2. By regulating chromatin state
[29], thereby influencing neighboring gene expression. In both cases, lncRNA can either silences or enhances targeted gene expression
[30]. The lncRNA Xist provides an ideal example of transcriptional silencing by spreading on a large portion of genes of one of the two X chromosomes
[31]. LncRNAs Gas6 and SWINGN are well-studied examples of enhancer and enhancer-associated lncRNAs, respectively, to promote the transcriptional activation of targeted proteins
[32].
1.2.2. Role in Post-Transcriptional Regulation
Regulation of post-transcriptional events comprises targeted protein modifications and their subcellular localization. The process includes mRNA splicing, processing, and nuclear export, with the help of RNA binding proteins (RBP). LncRNAs regulate gene expression at post-transcriptional levels by binding to RBP via RNA sequence motifs. LncRNA-mediated post-transcriptional regulation increases or decreases targeted mRNA and/or protein expression and their nuclear trafficking
[33]. Additionally, lncRNAs can alter mRNA stability and splicing in a context-dependent manner
[34]. The mRNA splicing regulation by lncRNAs involves either forming lncRNA-RNA hybrids with pre-mRNA or modifying the availability of splicing factors.
1.2.3. Role in Epigenetic Regulation
Many lncRNAs have been seen to regulate the epigenetic architecture of various proteins. These lncRNAs can either guide the epigenetic regulators to specific protein-coding loci or trigger the enhancer-promoter crosstalk. Their role as re-writers of chromatin folding (through chromatin modifying proteins such as methyltransferases and deacetylases) is documented in various studies
[35]. Recent advances in detecting lncRNA-chromatin association have lightened their role at the epigenetic level
[36]. Owing to their negatively charged surfaces, lncRNAs can neutralize positively charged histone proteins and de-compact the chromatin
[37].
2. Understanding the Role of LncRNAs in AGEs-Related DN
Studies have shown that lncRNAs play a significant role in AGEs-mediated metabolic malfunctions
[38]. So far, the involvement of lncRNAs-mediated AGEs-RAGE signaling in cancer, the immune system, and neurodegenerative diseases has been studied in detail
[39]. However, few studies have reported the role of lncRNAs in glycation-associated diabetic complications
[15][40][41][42][43] (
Table 2).
Table 2. LncRNAs involved in glycation/AGE-associated diabetic complications.
| LncRNA Involved |
Diabetic Complication |
Mode of Action |
References |
lncRNA
Arid2-IR |
Diabetic retinopathy |
Regulates oxidative stress, inflammatory responses, and endothelial cell dysfunction via interacting with Smad3 |
[40] |
| lncRNA MIAT |
Diabetic retinopathy |
AGE-induced HRPCs MIAT and CASP1 expressions increase, followed by the release of IL-1β, IL-18, and suppression of cell viability. |
[42] |
| lncRNA MEG3 |
Diabetic vascular diseases |
Upregulates in AGE-induced cells and suppresses cell viability and proliferation by modulating the MEG3/miR-193/p21 pathway |
[41] |
| lncRNA URIDS |
Diabetic Wound Healing |
Upregulates upon AGE induction. Regulates collagen production and deposition by targeting Plod1. It delays the wound healing process. |
[15] |
| lncRNA E330013P06 |
Inflammatory diabetes complications |
Increases inflammatory response upon AGE induction; by triggering pro-inflammatory gene. It also enhances foam cell formation. |
[43] |
2.1. LncRNA-Mediated RAGE Gene Expression and Signaling
The lncRNAs such as H9, MIAT, and MEG3 significantly regulate the pathogenesis of various diabetic complications. Recently, the role of lncRNA Arid2-IR in AGEs-induced retinal endothelial cells was also observed. By binding to Smad3, LncRNA Arid2-IR regulated levels of oxidative stress, inflammation, and apoptosis
[40]. Similarly, lncRNA HOTAIR regulated RAGE expression and inflammation in acute myocardium infarction-induced rat models
[44]. Likewise, the delayed process of diabetic wound healing in AGEs-induced fibroblast was associated with lncRNA URIDS
[15]. Overexpression and knockdown studies also confirmed the role of another lncRNA MVIH in the AGEs-RAGE signaling pathway responsible for tumor induction and progression in cancer
[45]. In AGEs-induced endothelial cells, lncRNA MEG3 regulated cell viability, proliferation, and apoptosis through the lncRNA MEG3/MiR-93/p21 mediated pathway during the onset of diabetic vascular diseases
[41]. In a cell-based study on hypoxia/reoxygenation-injured cardiomyocytes, lncRNA SNHG12 down-regulated RAGE, NF-κB expression, and pro-inflammatory responses
[46]. In a different context, the lncRNA TP73-AS1-mediated RAGE-HMGB1 signaling pathway upregulated NF-κB expression and pro-inflammatory cytokine levels
[12][47]. The above-discussed lncRNAs can be a promising biomarker to detect complications in AGEs-RAGE signaling-mediated metabolism.
2.2. LncRNAs That Regulate AGER Gene Expression and Signaling
Notably, the expression pattern of lncRNA AGER-1 correlates with AGER expression levels (r = 0.360,
p = 2.15 × 10
−18) possibly by binding and sponging miRNA-185. Animal-based studies confirmed these results with suppressed cell proliferation rate, migration, and colony-forming efficiency in nude mice
[48]. In a different context, lncRNA AGER-1 induced cell cycle arrest and promoted apoptosis, thereby inhibiting cell proliferation and migration efficiency of tumor cells in several types of cancer
[49]. Yet another study indicated the sponging effect of lncRNA AGER-1 for miR-182, an inhibitor of AGER1
[50]. Recently, a study showed that both AGER and its positive regulator lncRNA AGER-1 have the significant diagnostic potential for lung adenocarcinoma. Both AGER and lncRNA AGER-1 regulate apoptosis, cell migration, and antitumor responses
[51]. In summary, lncRNA AGER-1 can be a promising agent against AGEs signaling in metabolic disorders.