| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sorina Dinescu | + 3334 word(s) | 3334 | 2020-11-18 10:57:04 | | | |
| 2 | Rita Xu | -2125 word(s) | 1209 | 2020-11-27 03:18:44 | | |
Video Upload Options
Extracellular vesicles (EVs) are membranous structures produced in the endosomal system or generated by plasma membrane shedding, which have been identified as an important hallmark for intercellular communication. Among them, a particular category of EVs are the exosomes, which are nanovesicles of approximately 30-150 nm, produced in the endosomal pathway.
1. Introduction
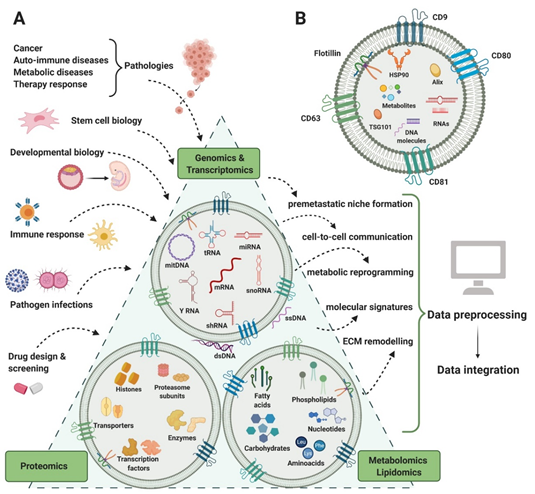
In the past 10 years, the scientific community’s perspective toward extracellular vesicles (EVs) has markedly focused on their role as mediators of intercellular communication, adding more insights to their known diversity and functions [1][2]. Underestimated in terms of heterogeneity, these membranous structures carry a diverse pool of functional biomolecules, most of them having unknown roles. So far, many attempts to screen these molecules or to construct reliable and reproducible molecular profiles of EV load have begun. Simultaneously, the rise of high-throughput applications in EV biology has enabled the collection of biological data at faster rates. These data are reported in various formats across a plethora of repositories, making searching and comparing results increasingly difficult. Computational methods can be used to provide integration strategies so that biological data can be observed in a system-wide manner, thus reflecting the elaborate interplay among biological variation at different levels of regulation [3].
The interest in multi-omics data integration emerges from the nature of a cell’s response to different conditions, which is best explained mechanistically when corroborating all omics levels [4]. Previous successful attempts of integrating multi-omics data have led to the characterization of informationally dense networks that accurately model human pathologies. By integrating genes and their known phenotypic profiles, groups of molecules that are involved in the same biological process (functional modules) have been identified [5]. Using a network-based data analysis approach, one could quantify how these modules are shared by similar diseases. According to how interconnected these modules are, disease-related genes having high association rates with a wide array of biomolecules may have a central role in the human interactome, often coding proteins acting as hubs in modulating systemic responses [6].
2. Biological Diversity of EVs
EVs are membranous structures produced in the endosomal system or generated by plasma membrane shedding. They are mainly found in biological fluids and are considered a viable way of cell-to-cell communication, with various implications in physiological and pathological processes [8][9]. So far, EVs have been isolated from a large variety of biological fluids such as plasma [10], urine [11], saliva [12], cerebrospinal fluid [13], breast milk [14], ascitic fluid [15], gastric juice [16], bile [17], sputum [18], bronchoalveolar lavage fluid [19], epididymal fluid [20], and tears [21]. EVs are a highly heterogeneous population, not only by means of size and biogenesis, but also composition and biomarkers, having specific DNA, RNA, protein, lipid, and metabolite cargo (figure 1). For a long period of time, size was the main characteristic used to classify EVs into exosomes and microvesicles (MVs), but contradictory results made it difficult and confusing, creating the need for alternative criteria [22][23][24][25]. As the EV biogenesis involves two main pathways that generate MVs and exosomes by membrane-trafficking processes, differentially expressed markers started being taken into consideration for a more accurate classification. Exosomes derive from the endosomal system, being synthesized as intraluminal vesicles within multivesicular bodies. Their sorting machineries are either dependent or independent of the endosomal sorting complex required for transport (ESCRT) proteins. MV generation is simpler, by plasma membrane shedding. In spite of the different packing mechanisms, the molecular profiles derived from both biogenesis processes overlap, making the MVs–exosomes dichotomy no longer useful [8][23][26] (Table 1).
Figure 1. EVs content in RNA, protein, lipid, and metabolites. Several areas of biology and medical research (input) generate complex genomics, transcriptomics, proteomics, lipidomics, and metabolomics analysis that can contribute to understanding several biological processes and functions (output).
Table 1. Evolution of extracellular vesicles (EV) dichotomous classification according to the accumulation of recent knowledge.
|
Historical Criteria |
Early Knowledge |
Current Knowledge |
|
Size |
MVs range between 100 and 1000 nm, while exosomes have dimeters smaller than 100 nm [23][27] |
Classification no longer in use, MVs can be smaller than 100 nm, exosomes have an upper limit based on endosomal size (up to 150 nm or larger); “small EVs” and “medium/large EVs” nomenclature is preferred [25] |
|
Protein content |
Different marker profiles due to biogenesis: GTP-binding proteins (ARF6), vesicle-associated membrane protein 3 (VAMP3), proteasomes, mitochondria-related proteins for MVs, transduction or scaffolding proteins (Syntenin 1), extracellular matrix, cell adhesion, receptor binding proteins and endosome-binding proteins (TSG101) for exosomes [24][28][29] |
No molecular markers that could characterize specifically each EV subtype, yet validation with three markers from three different classes is required in order to evaluate tissue specificity, lipid, or membrane-binding ability and purity [25] |
|
Lipid content |
Enriched contents according to the EV subtype: ceramides and sphingomyelins in MVs, cardiolipins in exosomes [29] |
Lipid ratios in EVs are not yet established [25]; more studies are needed in order to compare the lipid profiles of EVs with co-isolated lipoproteins and validate characteristic EV lipid contents such as lysoglycerophospholipids [30] |
|
Nucleic acid content |
DNA, mRNA, ncRNA, and especially miRNA in both MVs and exosomes; origin-specific miRNA profiles for exosomes [8][31] |
Confirmed specific incorporation of RNAs into subtypes of EVs [25] |
|
Isolation and purification methods |
Differential centrifugation or ultracentrifugation (10,000–20,000× g for MVs, 100,000–125,000× g for exosomes), size exclusion chromatography, immunoaffinity capture [32][33][34][35][36] |
No “golden standard” method to isolate and/or purify EVs, the choice is to be made based on the downstream applications, recovery, and specificity rates [24][25] |
EVs exert various biological roles, being involved in the immune response [37][38], regeneration [32], and as components of the extracellular matrix [39]. Early studies of EV function suggested antigen-presenting properties and the ability to stimulate T cell responses [34][40][41][42]. More recent studies indicate that EVs participate as well in autoimmune modulation processes in type 1 diabetes, as islets of Langerhans release EVs that are capable of activating peripheral blood mononuclear cells (PBMCs) [43]. In cancer, EVs are exchanged by cells within the tumor microenvironment in order to promote tumor growth and metastasis [44], vascularization [45], dormancy [46], chemoresistance [47], and metabolic reprogramming [48], mainly by transferring non-coding RNA molecules, such as miRNAs. Recently, EVs have been described as agents in modulating the spread of cancer cells toward their preferred metastases locations. Before invading other tissues and organs, tumors assure their growth by promoting the formation of a favorable micro-environment named the pre-metastatic niche (PMN) [49]. In colorectal and lung cancer, EVs enriched in small RNA species participate in activating pro-inflammatory processes through Toll-like receptor (TRL) pathways [50][51]. Other mechanisms that support metastasis are the mesothelial to mesenchymal transition and the promotion of angiogenesis through delivering miR-21-5p and miR-25-3p [52][53]. PMN formation as a result of cell-to-cell communication mediated by EVs was also observed in pancreatic, ovarian, and lung cancer [54][55][56].
As EVs play an important role in tumor growth and metastasis, it is fair to consider that they may represent therapeutic targets as well. For this matter, three strategies that might alleviate the EV contribution to tumor development were postulated: the elimination of circulating EVs based on specific surface markers, the inhibition of EV release, and the impairment of EV absorption [57]. For example, targeting the gene for the autophagy related 5 (ATG5) protein that stimulates in vivo metastasis through EV production might have promising results in holding tumor progression [58]. Drug repurposing is another promising alternative, as FDA-approved compounds such as the antibiotic sulfisoxazole impair EV secretion in breast cancer cells [59]. Taken together, EVs are of great interest not only for their crucial implications in physiological and pathological processes but also for their clinical promise.
References
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Mol. Life Sci. 2011, 68, 2667–2688, doi:10.1007/s00018-011-0689-3.
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. Extracell. Vesicles 2019, 8, 1648167, doi:10.1080/20013078.2019.1648167.
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Rev. Genet. 2015, 16, 85–97, doi:10.1038/nrg3868.
- Sass, S.; Buettner, F.; Mueller, N.S.; Theis, F.J. A modular framework for gene set analysis integrating multilevel omics data. Nucleic Acids Res. 2013, 41, 9622, doi:10.1093/NAR/GKT752.
- Hartwell, L.H.; Hopfield, J.J.; Leibler, S.; Murray, A.W. From molecular to modular cell biology. Nature 1999, 402, C47-52, doi:10.1038/35011540.
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Natl. Acad. Sci. 2007, 104, 8685–8690, doi:10.1073/PNAS.0701361104.
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335, doi:10.1038/nature15756.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Rev. Mol. Cell Biol. 2018, 19, 213–228, doi:10.1038/nrm.2017.125.
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. Extracell. Vesicles 2015, 4, 27066, doi:10.3402/jev.v4.27066.
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Immunol. 2005, 17, 879–887, doi:10.1093/intimm/dxh267.
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Natl. Acad. Sci. U. S. A. 2004, 101, 13368–13373, doi:10.1073/pnas.0403453101.
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Pharm. Bull. 2008, 31, 1059–1062, doi:10.1248/bpb.31.1059.
- Harrington, M.G.; Fonteh, A.N.; Oborina, E.; Liao, P.; Cowan, R.P.; McComb, G.; Chavez, J.N.; Rush, J.; Biringer, R.G.; Hühmer, A.F. The morphology and biochemistry of nanostructures provide evidence for synthesis and signaling functions in human cerebrospinal fluid. Cerebrospinal Fluid Res. 2009, 6, 10, doi:10.1186/1743-8454-6-10.
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. Immunol. 2007, 179, 1969–1978, doi:10.4049/jimmunol.179.3.1969.
- Gutwein, P.; Stoeck, A.; Riedle, S.; Gast, D.; Runz, S.; Condon, T.P.; Marmé, A.; Phong, M.-C.; Linderkamp, O.; Skorokhod, A.; et al. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Cancer Res. 2005, 11, 2492–2501, doi:10.1158/1078-0432.CCR-04-1688.
- Yamamoto, H.; Watanabe, Y.; Oikawa, R.; Morita, R.; Yoshida, Y.; Maehata, T.; Yasuda, H.; Itoh, F. BARHL2 Methylation Using Gastric Wash DNA or Gastric Juice Exosomal DNA is a Useful Marker For Early Detection of Gastric Cancer in an H. pylori-Independent Manner. Transl. Gastroenterol. 2016, 7, e184, doi:10.1038/ctg.2016.40.
- Witek, R.P.; Yang, L.; Liu, R.; Jung, Y.; Omenetti, A.; Syn, W.-K.; Choi, S.S.; Cheong, Y.; Fearing, C.M.; Agboola, K.M.; et al. Liver cell-derived microparticles activate hedgehog signaling and alter gene expression in hepatic endothelial cells. Gastroenterology 2009, 136, 320–330.e2, doi:10.1053/j.gastro.2008.09.066.
- Porro, C.; Lepore, S.; Trotta, T.; Castellani, S.; Ratclif, L.; Battaglino, A.; Di Gioia, S.; Martínez, M.C.; Conese, M.; Maffione, A.B. Isolation and characterization of microparticles in sputum from cystic fibrosis patients. Res. 2010, 11, 94, doi:10.1186/1465-9921-11-94.
- Wahlund, C.J.E.; Eklund, A.; Grunewald, J.; Gabrielsson, S. Pulmonary Extracellular Vesicles as Mediators of Local and Systemic Inflammation. cell Dev. Biol. 2017, 5, 39, doi:10.3389/fcell.2017.00039.
- Gatti, J.-L.; Métayer, S.; Belghazi, M.; Dacheux, F.; Dacheux, J.-L. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Reprod. 2005, 72, 1452–1465, doi:10.1095/biolreprod.104.036426.
- Grigor’eva, A.E.; Tamkovich, S.N.; Eremina, A. V; Tupikin, A.E.; Kabilov, M.R.; Chernykh, V. V; Vlassov, V. V; Laktionov, P.P.; Ryabchikova, E.I. [Characteristics of exosomes andmicroparticles discovered in human tears]. Khim. 2016, 62, 99–106, doi:10.18097/PBMC20166201099.
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Tigges, J.; Toxavidis, V.; Ericsson, M.; Distel, R.J.; Ivanov, A.R.; Skog, J.; Kuo, W.P. Alternative Methods for Characterization of Extracellular Vesicles. Physiol. 2012, 3, 354, doi:10.3389/fphys.2012.00354.
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Mol. Neurobiol. 2016, 36, 301–312, doi:10.1007/s10571-016-0366-z.
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I. V; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Extracell. vesicles 2014, 3, 26913, doi:10.3402/jev.v3.26913.
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Extracell. Vesicles 2018, 7, doi:10.1080/20013078.2018.1535750.
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727, doi:10.3390/cells8070727.
- Choi, D.-S.; Kim, D.-K.; Kim, Y.-K.; Gho, Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571, doi:10.1002/pmic.201200329.
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Neurooncol. 2013, 113, 1–11, doi:10.1007/s11060-013-1084-8.
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. Extracell. Vesicles 2016, 5, 32570, doi:10.3402/jev.v5.32570.
- Sun, Y.; Saito, K.; Saito, Y. Lipid Profile Characterization and Lipoprotein Comparison of Extracellular Vesicles from Human Plasma and Serum. Metabolites 2019, 9, doi:10.3390/metabo9110259.
- Ji, H.; Chen, M.; Greening, D.W.; He, W.; Rai, A.; Zhang, W.; Simpson, R.J. Deep Sequencing of RNA from Three Different Extracellular Vesicle (EV) Subtypes Released from the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct Mirna-Enrichment Signatures. PLoS One 2014, 9, e110314, doi:10.1371/journal.pone.0110314.
- Ren, S.; Chen, J.; Duscher, D.; Liu, Y.; Guo, G.; Kang, Y.; Xiong, H.; Zhan, P.; Wang, Y.; Wang, C.; et al. Microvesicles from human adipose stem cells promote wound healing by optimizing cellular functions via AKT and ERK signaling pathways. Stem Cell Res. Ther. 2019, 10, 47, doi:10.1186/s13287-019-1152-x.
- Leblanc, P.; Arellano-Anaya, Z.E.; Bernard, E.; Gallay, L.; Provansal, M.; Lehmann, S.; Schaeffer, L.; Raposo, G.; Vilette, D. Isolation of Exosomes and Microvesicles from Cell Culture Systems to Study Prion Transmission. In Exosomes and Miceovesicles; Hill, A., Ed.; Humana Press, New York, NY, 2017; pp. 153–176.
- Pérez-González, R.; Gauthier, S.A.; Kumar, A.; Saito, M.; Saito, M.; Levy, E. A Method for Isolation of Extracellular Vesicles and Characterization of Exosomes from Brain Extracellular Space. In Exosomes and Microvesicles; Hill, A., Ed.; Humana Press, New York, NY, 2017; pp. 139–151.
- Lane, R.E.; Korbie, D.; Trau, M.; Hill, M.M. Purification Protocols for Extracellular Vesicles. In Extracellular Vesicles; Kuo, W., Jia, S., Eds.; Humana Press, New York, NY, 2017; pp. 111–130.
- Huang, T.; He, J. Characterization of Extracellular Vesicles by Size-Exclusion High-Performance Liquid Chromatography (HPLC). In Extracellular Vesicles; Humana Press: New York, NY, USA, 2017; pp. 191–199.
- Mallegol, J.; Van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.K.; Raposo, G.; Cerf–Bensussan, N.; Heyman, M. T84-Intestinal Epithelial Exosomes Bear MHC Class II/Peptide Complexes Potentiating Antigen Presentation by Dendritic Cells. Gastroenterology 2007, 132, 1866–1876, doi:10.1053/J.GASTRO.2007.02.043.
- Rossaint, J.; Kühne, K.; Skupski, J.; Van Aken, H.; Looney, M.R.; Hidalgo, A.; Zarbock, A. Directed transport of neutrophil-derived extracellular vesicles enables platelet-mediated innate immune response. Commun. 2016, 7, 13464, doi:10.1038/ncomms13464.
- Rilla, K.; Mustonen, A.-M.; Arasu, U.T.; Härkönen, K.; Matilainen, J.; Nieminen, P. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol. 2019, 75–76, 201–219, doi:10.1016/J.MATBIO.2017.10.003.
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C. V; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. Exp. Med. 1996, 183, 1161–1172, doi:10.1084/jem.183.3.1161.
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes. Med. 1998, 4, 594–600, doi:10.1038/nm0598-594.
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell–derived exosomes. Immunol. 2002, 3, 1156–1162, doi:10.1038/ni854.
- Rutman, A.K.; Negi, S.; Gasparrini, M.; Hasilo, C.P.; Tchervenkov, J.; Paraskevas, S. Immune Response to Extracellular Vesicles From Human Islets of Langerhans in Patients With Type 1 Diabetes. Endocrinology 2018, 159, 3834–3847, doi:10.1210/en.2018-00649.
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated Polarization of Tumor-Associated Macrophages by miR-145 via Colorectal Cancer-Derived Extracellular Vesicles. Immunol. 2017, 199, 1505–1515, doi:10.4049/jimmunol.1700167.
- Ludwig, N.; Yerneni, S.S.; Razzo, B.M.; Whiteside, T.L. Exosomes from HNSCC Promote Angiogenesis through Reprogramming of Endothelial Cells. Cancer Res. 2018, 16, 1798–1808, doi:10.1158/1541-7786.MCR-18-0358.
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell–Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844, doi:10.1158/0008-5472.CAN-16-1092.
- Santos, J.C.; Ribeiro, M.L.; Sarian, L.O.; Ortega, M.M.; Derchain, S.F. Exosomes-mediate microRNAs transfer in breast cancer chemoresistance regulation. J. Cancer Res. 2016, 6, 2129.
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation. Cancer 2018, 17, 97, doi:10.1186/s12943-018-0846-5.
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: organ-specific homes for metastases. Rev. Cancer 2017, 17, 302–317, doi:10.1038/nrc.2017.6.
- Shao, Y.; Chen, T.; Zheng, X.; Yang, S.; Xu, K.; Chen, X.; Xu, F.; Wang, L.; Shen, Y.; Wang, T.; et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018, 39, 1368–1379, doi:10.1093/carcin/bgy115.
- Liu, Y.; Gu, Y.; Han, Y.; Zhang, Q.; Jiang, Z.; Zhang, X.; Huang, B.; Xu, X.; Zheng, J.; Cao, X. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256, doi:10.1016/j.ccell.2016.06.021.
- Li, Q.; Li, B.; Li, Q.; Wei, S.; He, Z.; Huang, X.; Wang, L.; Xia, Y.; Xu, Z.; Li, Z.; et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018, 9, 854, doi:10.1038/s41419-018-0928-8.
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Commun. 2018, 9, 5395, doi:10.1038/s41467-018-07810-w.
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Cell Biol. 2015, 17, 816–826, doi:10.1038/ncb3169.
- Feng, W.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Cancer 2019, 18, 124, doi:10.1186/s12943-019-1049-4.
- Kong, J.; Tian, H.; Zhang, F.; Zhang, Z.; Li, J.; Liu, X.; Li, X.; Liu, J.; Li, X.; Jin, D.; et al. Extracellular vesicles of carcinoma-associated fibroblasts creates a pre-metastatic niche in the lung through activating fibroblasts. Cancer 2019, 18, 175, doi:10.1186/s12943-019-1101-4.
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. J. Physiol. Physiol. 2020, 318, C29–C39, doi:10.1152/ajpcell.00280.2019.
- Guo, H.; Chitiprolu, M.; Roncevic, L.; Javalet, C.; Hemming, F.J.; Trung, M.T.; Meng, L.; Latreille, E.; Tanese de Souza, C.; McCulloch, D.; et al. Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Cell 2017, 43, 716–730, doi:10.1016/j.devcel.2017.11.018.
- Im, E.-J.; Lee, C.-H.; Moon, P.-G.; Rangaswamy, G.G.; Lee, B.; Lee, J.M.; Lee, J.-C.; Jee, J.-G.; Bae, J.-S.; Kwon, T.-K.; et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Commun. 2019, 10, 1387, doi:10.1038/s41467-019-09387-4.