
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Seitaro Nomura | + 1361 word(s) | 1361 | 2020-11-12 09:55:42 | | | |
| 2 | Dean Liu | -232 word(s) | 1129 | 2020-11-19 08:38:22 | | |
Video Upload Options
Single-cell RNA sequencing (scRNA-seq) technology is a powerful, rapidly developing tool for characterizing individual cells and elucidating biological mechanisms at the cellular level. Cardiovascular disease is one of the major causes of death worldwide and its precise pathology remains unclear. scRNA-seq has provided many novel insights into both healthy and pathological hearts. In this review, we summarize the various scRNA-seq platforms and describe the molecular mechanisms of cardiovascular development and disease revealed by scRNA-seq analysis. We then describe the latest technological advances in scRNA-seq. Finally, we discuss how to translate basic research into clinical medicine using scRNA-seq technology.
1. Introduction
To understand the phenomenon of life, it is important to elucidate the biological mechanisms of the cells that constitute living organisms. RNA is essential to biological processes in cells, and transcriptomes provide critical information directly associated with cell phenotypes. Single-cell RNA sequencing (scRNA-seq) is a powerful tool for characterizing individual cells. The conventional technique of bulk RNA-seq measures the average gene expression across cells in a sample and identifies differences between sample conditions, whereas scRNA-seq measures the gene expression of individual cells and can identify differences between cells in one or more samples. Although cells are traditionally characterized morphologically or by molecules unique to each cell type, scRNA-seq facilitates automatic classification of cells via clustering of transcriptomes and can identify heterogeneous cell types and molecular states even in a group that have been considered to consist of only one cell type [1].
Cardiovascular disease is common worldwide. One in a hundred children have congenital heart disease, such as ventricular septal defect and tetralogy of Fallot, resulting from an abnormality during the heart development process. For adults, cardiovascular disease is one of the major causes of death worldwide and includes cardiomyopathy, ischemic heart disease, and valvular disease, which can lead to heart failure and even sudden death. The precise pathology remains to be elucidated because of the complexity of interactions between cells and molecules not only within the heart but also across other organs.
The heart is exposed to various types of stress. Cardiomyocytes (CMs) have an abundant supply of mitochondria, which not only allows them to produce energy for contraction but also generates reactive oxygen species and leads to cellular toxicity [2]. The heart constantly contracts and is under high pressure from the blood, especially in the left ventricle, which can cause mechanical stress and hypertrophy [3]. Several molecules involved in the renin–angiotensin system and the sympathetic nervous system and cytokines affect the function of the heart.
In past decades, conventional molecular biological studies have partly revealed the pathological mechanism underlying cardiovascular disease, but further studies are warranted. The development of scRNA-seq techniques could provide new insights into both healthy and pathological hearts.
The content of this review is summarized in Figure 1. We first introduce the scRNA-seq platforms appropriate for each cell type. Next, we describe the molecular mechanisms of cardiovascular development and disease revealed by scRNA-seq analysis. Then, we present the latest technological advances in scRNA-seq. Finally, we discuss how to translate basic research into clinical medicine using scRNA-seq technology.
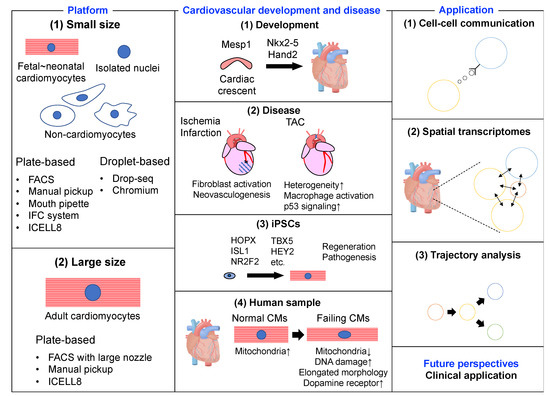
Figure 1. Summary figure of single-cell RNA-sequencing in the heart.
2. scRNA-seq Platforms
2.1. Platforms for Small Cells
Dispensing tissues into a single cell is an important process in scRNA-seq analysis. Fluorescence-activated cell sorting (FACS) is conventionally used for single-cell sorting [4] and utilizes scatter and fluorescence signals to sort cells into 96- or 384-well plates. The use of fluorescence signals enables the filtering of dead cells and the selection of cells of interest. Because of the size of the nozzle, cells less than 50 μm in diameter can be sorted with minimal damage, but it is difficult to sort adult CMs due to their large size (>100 μm in diameter along the major axis). Another option is to use the integrated fluidic circuit (IFC) system (e.g., Fluidigm C1), which isolates single cells into individual reaction chambers in a more automated manner. Cells are automatically lysed, and cDNA libraries are quickly prepared for sequencing. IFC captures cells less than 25 μm in diameter and processes up to 800 cells. FACS and IFC are plate-based platforms, and thus the number of cells that can be sorted and analyzed is limited.
Both Drop-seq [5] and Chromium (10× Genomics), which is a commercial droplet-based platform, allow for the rapid profiling of thousands of individual cells by encapsulating them in tiny droplets, adding barcodes to each cell’s RNAs, and sequencing them together. These approaches utilize a microfluidics device to perform droplet separation; therefore, the size of cells that can be analyzed is generally less than 30 μm in diameter.
The droplet-based system is superior to the plate-based system in terms of throughput, but the former tends to be less effective at detecting genes due to its low capture rate of mRNA molecules and it has a higher tendency to generate doublets. Most studies using a droplet system detected around 500–1500 genes, whereas those using a plate-based system detected around 1000–4000 genes. It is important to select methods according to how many cells will be analyzed and how many genes are expected to be detected.
These plate-based (FACS and IFC system) and droplet-based platforms (Drop-seq and Chromium) are suitable for scRNA-seq of normal and small cells. Although adult CMs in mice and humans are relatively large, embryonic and neonatal CMs can be analyzed using such systems. In addition, non-CMs, such as fibroblasts, endothelial cells, and macrophages, and nuclei extracted from adult CMs are small enough to be analyzed using these systems.
2.2. Platforms for Large Cells
The channel diameter of the conventional nozzle in FACS, IFC, and droplet-based systems is too narrow for adult CMs to pass through without damage. Gladka et al. used FACS with a larger nozzle size (130 μm) to perform scRNA-seq of adult murine CMs [6]. ICELL8 is another promising platform that uses a large-bore nozzle dispenser to distribute single cells from diluted cell suspensions into 5184 nanowells. ICELL8 also has an imaging system that visualizes all nanowells. Using fluorescence signals of the imaging system makes it possible to differentiate live cells from dead cells, or wells containing a single cell from wells containing no or multiple cells, generating 1000–1500 single-cell transcriptomes. Manual pick-up and mouth pipettes are also utilized in the scRNA-seq of adult CMs because they are gentler on cells in terms of stress compared with FACS. Using these strategies, we can use a microscope to choose the cardiomyocytes with the best shape for sequencing. However, some training is necessary for manual sorting of individual cells and the throughput is lower.
There are few platforms for scRNA-seq of adult CMs, and given that all of them are plate-based systems, the number of cells they can analyze is limited. Therefore, some researchers have used nuclei extracted from adult CMs, rather than intact CMs, in order to utilize the droplet-based system.
References
- Lafzi, A.; Moutinho, C.; Picelli, S.; Heyn, H. Tutorial: Guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 2018, 13, 2742–2757.
- Kornfeld, O.S.; Hwang, S.; Disatnik, M.H.; Chen, C.H.; Qvit, N.; Mochly-Rosen, D. Mitochondrial reactive oxygen species at the heart of the matter: New therapeutic approaches for cardiovascular diseases. Circ. Res. 2015, 116, 1783–1799.
- Komuro, I. Molecular mechanism of mechanical stress-induced cardiac hypertrophy. Jpn. Heart J. 2000, 41, 117–129.
- Ibrahim, S.F.; van den Engh, G. Flow cytometry and cell sorting. Adv. Biochem. Eng. Biotechnol. 2007, 106, 19–39.
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214.
- Gladka, M.M.; Molenaar, B.; de Ruiter, H.; van der Elst, S.; Tsui, H.; Versteeg, D.; Lacraz, G.P.A.; Huibers, M.M.H.; van Oudenaarden, A.; van Rooij, E. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation 2018, 138, 166–180.




