
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dean Liu | -- | 1949 | 2022-10-17 01:44:45 |
Video Upload Options
GeneCards is a database of human genes that provides genomic, proteomic, transcriptomic, genetic and functional information on all known and predicted human genes. It is being developed and maintained by the Crown Human Genome Center at the Weizmann Institute of Science. The database aims at providing a quick overview of the current available biomedical information about the searched gene, including the human genes, the encoded proteins, and the relevant diseases. The GeneCards database provides access to free Web resources about more than 7000 all known human genes that integrated from >90 data resources, such as HGNC, Ensembl, and NCBI. The core gene list is based on approved gene symbols published by the HUGO Gene Nomenclature Committee (HGNC). The information is carefully gathered and selected from these databases by its engine. If the search does not return any results, this database will give several suggestions to help users accomplish their search depending on the type of query and offer direct links to other databases’ search engine. Over time, the GeneCards database has developed a suite of tools (GeneDecks, GeneLoc, GeneALaCart) that has more specialised capability. Since 1998, the GeneCards database has been widely used by bioinformatics, genomics and medical communities for more than 15 years.
1. History
Since the 1980s, sequence information has become increasingly abundant; subsequently many laboratories realized this and began to store such information in central repositories-the primary database.[1] However, the information provided by the primary sequence databases (lower level databases) focus on different aspects. To gather these scattered data, the Weizmann Institute of Science's Crown Human Genome Centre developed a database called ‘GeneCards’ in 1997. This database mainly dealt with human genome information, human genes, the encoded proteins’ functions, and related diseases, though it has expanded since that time.[2]
2. Growth
Initially, the GeneCards database had two main features: delivery of integrated biomedical information for a gene in ‘card’ format, and a text-based search engine. Since 1998, the database has integrated more data resources and data types, such as protein expression and gene network information. It has also improved the speed and sophistication of the search engine, and expanded from a gene-centric dogma to contain gene-set analyses. Version 3 of the database gathers information from more than 90 database resources based on a consolidated gene list. It has also added a suite of GeneCards tools which focus on more specific purposes. "GeneNote and GeneAnnot for transcriptome analyses, GeneLoc for genomic locations and markers, GeneALaCart for batch queries and GeneDecks for finding functional partners and for gene set distillations.". The database updates on a 3-year cycle of planning, implementation, development, semi-automated quality assurance, and deployment. Technologies used include Eclipse, Apache, Perl, XML, PHP, Propel, Java, R and MySQL.[3][4]
3. Ongoing GeneCards Expansions[3]
- Animal models
- Tissue proteomics profiling
- RNA genes
- Gene and protein identifier mapping
- Online analytical processing (OLAP)
4. Availability
GeneCards can be freely accessed by non-profit institution for educational and research purpose at https://www.genecards.org/ and academic mirror sites. Commercial usage requires a license.
5. GeneCards Suite
5.1. GeneDecks
GeneDecks is a novel analysis tool to identify similar or partner genes, which provides a similarity metric by highlighting shared descriptors between genes, based on GeneCards’ unique wealth of combinatorial annotations of human genes.
- Annotation combinatory: Using GeneDecks, one can get a set of similar genes for a particular gene with a selected combinatorial annotation. The summary table result in ranking the different level of similarity between the identified genes and the probe gene.
- Annotation unification: Different data sources often offer annotations with heterogeneous naming system. Annotation unification of GeneDecks is based on the similarity in GeneCards gene-content space detection algorithms.
- Partner hunting: In GeneDecks's Partner Hunter, users give a query gene, and the system seeks similar genes based on combinatorial similarity of weighted attributes.
- Set distillation: In Set distiller, users give a set of genes, and the system ranks attributes by their degree of sharing within a given gene set. Like Partner Hunter, it enables sophisticated investigation of a variety of gene sets, of diverse origins, for discovering and elucidating relevant biological patterns, thus enhancing systematic genomics and systems biology scrutiny.[4][5][6]
5.1. GeneALaCart
GeneALaCart is a gene-set-orientated batch-querying engine based on the popular GeneCards database. It allows retrieval of information about multiple genes in a batch query.[3][7]
5.2. GeneLoc
The GeneLoc suit member presents an integrated human chromosome map, which is very important for designing a custom-made capture chip, based on data integrated by the GeneLoc algorithm. GeneLoc includes further links to GeneCards, NCBI's Human Genome Sequencing, UniGene, and mapping resources.[3][8]
6. Usage
6.1. Search
Firstly, enter a search term into the blank on the homepages. Searching methods include Keywords, Symbol only, Symbol/Alias/Identifier and Symbol/Alias.[9] The default search option is searching by keywords. When a user searches by keywords, MicroCard and MiniCard are shown. However, when a user searches by Symbol only, they will be directed to GeneCard.[10] Searches may be furthered by clicking on advanced search, where a user can choose section, category, GIFtS, Symbol Source and gene sets directly. Sections include Aliases & Descriptions, Disorders, Drugs & Compounds, Expression in Human Tissues, Function, Genomic Location, Genomic Variants, Orthologs, Paralogs, Pathways & Interactions, Protein Domains/Families, Proteins, Publications, Summaries and Transcripts. The default option is searching for all sections.[9] Categories include Protein-coding, Pseudogenes, RNA genes Genetic Loci, Gene clusters and Uncategorized. The default option is searching for all categories.[9] GIFtS is the GeneCards Inferred Functionality Scores, which gives objective numbers to show the knowledge level about the functionality of human genes. It includes High, Medium, Low, and custom range.[11][12] Symbol Sources include HGNC (HUGO Gene Nomenclature Committee), EntrezGene (gene-centered information at NCBI), Ensembl, GeneCards RNA genes, CroW21 and so on.[9]
Moreover, the user can choose to search for All GeneCards or Within Gene Subset, which would be more specific and with priority.
Secondly, the search result page shows all relevant minicards. Symbol, Description, Category, GIFtS, GC id and Score are displayed on the page.[9] A user may click on the plus button for each of the mini-cards to open the minicard. Also, the user can click directly on the symbol to see the details of a particular GeneCard.
6.2. GeneCards Content[9]
For a particular GeneCard (example: GeneCard for TGFB1), it is consist of the following contents.
- Header: The header is made up of gene's symbol, category (i.e. protein-coding), GIFtS(i.e. 74) and GCID(GC19M041837). Different categories have different colors to express: protein-coding, pseudogene, RNA gene, gene cluster, genetic locus, and uncategorized. The background indicates the symbol sources: HGNC Approved Genes, EntrezGene Database, Ensembl Gene Database, or GeneCards Generated Genes.
- Aliases: Aliases, as its name indicates, shows synonyms and aliases of the gene according to diverse sources such as HGNC. The right column displays how the aliases associated with the resources and gives previous GC identifiers.
- Summaries: The left column is the same with the one in the Aliases, which shows the sources. The right column here gives brief summary on gene's function, localization and effect on phenotype from various sources.
- Genomic Views: In addition to sources, this section gives reference DNA sequence, regulatory elements, epigenetics, chromosome band and genomic location of different sources. The red line on the image indicates the GeneLoc integrated location. In particular, if the GeneLoc integrated location is different from the location in Entrez Gene, it is shown in green; Blue is appeared when the GeneLoc integrated location differs from the location in Ensembl. Addition details can be accessed through the links in the section.
- Proteins: This section presents annotated information of genes, including recommended name, size, subunit, subcellular location and secondary accessions. Also, post-translational modifications, protein expression data, REF SEQ proteins, ENSEMBL proteins, Reactome Protein details, Human Recombinant Protein Products, Gene Ontology, Antibody Products and Assay Products are introduced.
- Protein Domains/Families: This section shows annotated information of protein domains and families.
- Function: The function section describes gene function, including: Human phenotypes, bound Targets, shRNA for human and/or mouse/rat, miRNA Gene Targets, RNAi products, microRNA for human and/or mouse/rat orthologs, Gene Editing, Clones, Cell Lines, Animal models, in situ hybridization assays.
- Pathways & Interactions: This section shows unified GeneCards pathways and interactions that are from different sources. Unified GeneCards pathways are collected into super-pathways, which displays the connection between different pathways. Interaction shows interactant and interaction details.
- Drugs & Compounds: This section connects GeneCards with drugs and compounds. TOCRIS compounds show compound, action and CAS number. DrugBank compound gives compound, synonyms, CAS number (Chemical Abstracts Registry number), type (transporter/target/carrier/enzyme), actions and PubMed IDs. HMDB and Novoseek show the relationships of chemical compounds, which includes compound, synonyms, CAS number and PubMed IDs (articles related to the compound). BitterDB displays compound, CAS number and SMILES (Simplified Molecular Input Line Entry Specification). PharmGKB gives drug/compound and its annotation.
- Transcripts: This section is consist of reference sequence mRNAs, Unigene Cluster and representative Sequence, miRNA products, inhib.RNA products, Clone products, primer products and additional mRNA sequence. Also, the user can gain exon structure from GeneLoc.
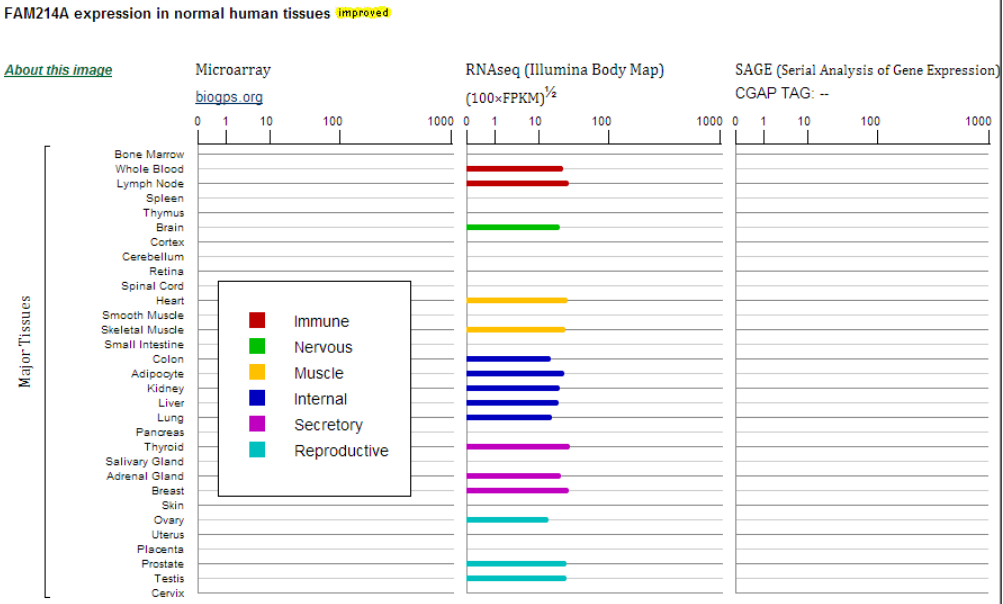
- Expression: The left column shows the resources of the data. Expression images and data, similar genes, PCR arrays, primers for human and in situ hybridization assays are included in this section.
- Orthologs: This section gives orthologs for a particular gene from numbers of species. The table displays the corresponding organism, taxonomic classification, gene, description, human similarity, orthology type and details. It's connected to ENSEMBL Gene Tree, TreeFam Gene Tree, and Aminode.[13]
- Paralogs: This section displays paralogs and pseudogenes for a particular gene.
- Genomic Variants: The genomic variants show the result of NCBI SNPs/Variants, HapMap linkage disequilibrium report, structural variations, human gene mutation database(HGMD), QIAGEN SeqTarget long-range PCR primers in human, mouse &rat and SABiosciences cancer mutation PCR arrays. The table in this section shows SNP ID, Valid, Clinical significance, Chr pos, Sequence for genomic data, AAChg, Type and More for transcription related data, Allele freq, Pop, Total sample and More for Allele Frequencies. For Valid, the different character represents different validation methods. ‘C’ means by-cluster; ‘A’ is by-2hit-2allele; ‘F’ is by-frequency; ‘H’ is by-hapmap and ‘O’ is by-other-pop. Clinical significance can be one of the following: non-pathogenic, pathogenic, drug-response, histocompatibility, probable-non-pathogenic, probable-pathogenic, untested, unknown and other. Type should be one of these: nonsynon, syn, cds, spl, utr, int, exc, loc, stg, ds500, spa, spd, us2k, us5k, PupaSUITE Designations.
- Disorders/Diseases: Shows disorders/diseases associated with the gene.
- Publications: Displays publications associated with the gene.
- External Searches: Searches more information in PubMed, OMIM and NCBI.
- Genome Databases: Other Databases, and specialized Databases.
- Intellectual Property: This section gives patent information and licensable technologies.
- Products
7. Applications
GeneCards is used widely in the biological and biomedical fields. For example, S.H. Shah extracted data of early-onset coronary artery disease from GeneCards to identify genes that contributes to the disease. Chromosome 3q13, 1q25 etc. are confirmed to take effects and this paper further discussed the relationship between morbid genes and serum lipoproteins with the help of GeneCard.[14]
Another example is a research study on synthetic lethality in cancer. Synthetic lethality appears when a mutation in a single gene has no effect on the function of a cell but a mutation in an additional gene leads to cell death. This study aimed to find novel methods of treating cancer through blocking the lethality of drugs. GeneCards was used when comparing data of a given target gene with all possible genes. In this process, the annotation sharing score was calculated using GeneDecks Partner Hunter (now called Genes Like Me) to give paralogy. Inactivation targets were be extracted after the microarray experiments of resistant and non-resistant neuroblastoma cell lines.[3]
References
- Attwood, Teresa K.; Parry-Smith, David J. (1999). Introduction to Bioinformatics. Harlow Longman.
- "GeneCards: integrating information about genes, proteins and diseases". Trends in Genetics 13 (4): 163. April 1997. doi:10.1016/S0168-9525(97)01103-7. PMID 9097728. https://dx.doi.org/10.1016%2FS0168-9525%2897%2901103-7
- "In-silico human genomics with GeneCards". Human Genomics 5 (6): 709–17. October 2011. doi:10.1186/1479-7364-5-6-709. PMID 22155609. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3525253
- "GeneCards Version 3: the human gene integrator". Database 2010: baq020. August 2010. doi:10.1093/database/baq020. PMID 20689021. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2938269
- "GeneDecks". GeneCards. http://genecards.org/index.php?path=/GeneDecks/aboutGeneDecks.
- "GeneDecks: paralog hunting and gene-set distillation with GeneCards annotation". Omics 13 (6): 477–87. December 2009. doi:10.1089/omi.2009.0069. PMID 20001862. https://dx.doi.org/10.1089%2Fomi.2009.0069
- "GeneALaCart". GeneCards. http://genecards.org/BatchQueries/index.php.
- "GeneLoc". GeneCards. http://genecards.weizmann.ac.il/geneloc/index.shtml.
- "GeneCards". GeneCards. http://genecards.org/.
- "Bioinformatics approach to extract information from genes". Journal of Diabetes in Developing Countries 26 (4): 149–51. 2006. doi:10.4103/0973-3930.33179. https://dx.doi.org/10.4103%2F0973-3930.33179
- "GIFtS: annotation landscape analysis with GeneCards". BMC Bioinformatics 10 (1): 348. October 2009. doi:10.1186/1471-2105-10-348. PMID 19852797. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2774327
- "GeneAnnot: interfacing GeneCards with high-throughput gene expression compendia". Briefings in Bioinformatics 4 (4): 349–60. December 2003. doi:10.1093/bib/4.4.349. PMID 14725348. https://dx.doi.org/10.1093%2Fbib%2F4.4.349
- "Aminode: Identification of Evolutionary Constraints in the Human Proteome". Scientific Reports 8 (1): 1357. January 2018. doi:10.1038/s41598-018-19744-w. PMID 29358731. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5778061
- "Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q". Annals of Human Genetics 70 (Pt 6): 738–48. November 2006. doi:10.1111/j.1469-1809.2006.00288.x. PMID 17044848. https://dx.doi.org/10.1111%2Fj.1469-1809.2006.00288.x





