
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Teresa Semedo-Lemsaddek | -- | 2914 | 2022-10-14 11:17:25 | | | |
| 2 | Jessie Wu | Meta information modification | 2914 | 2022-10-17 03:02:40 | | | | |
| 3 | Jessie Wu | Meta information modification | 2914 | 2022-10-17 03:07:14 | | | | |
| 4 | Jessie Wu | Meta information modification | 2914 | 2022-10-17 03:07:45 | | | | |
| 5 | Jessie Wu | Meta information modification | 2914 | 2022-10-17 08:09:09 | | |
Video Upload Options
The family Enterobacteriaceae is a large, heterogeneous group of Gram-negative bacteria, which includes strains that naturally inhabit the gastrointestinal tract (GIT) of animals and humans. Being considered normal commensal members of the GIT microbiota, these microbes can also live and multiply in food environments. Additionally, Enterobacteriaceae are acknowledged as indicators of food production hygiene, preservation, and storage, often being used as indicators of food quality and safety. Additionally, Salmonella spp., Yersinia enterocolitica, pathogenic Escherichia coli, including Escherichia coli O157:H7, and Shigella spp., among others, are important foodborne pathogens. Moreover, some members of this group, namely Citrobacter, Enterobacter, Erwinia, Klebsiella, Kluyvera, Pantoea, and Serratia, have been described both as harboring plant growth-promoting characteristics and attaining pathogenicity potential.
1. Introduction
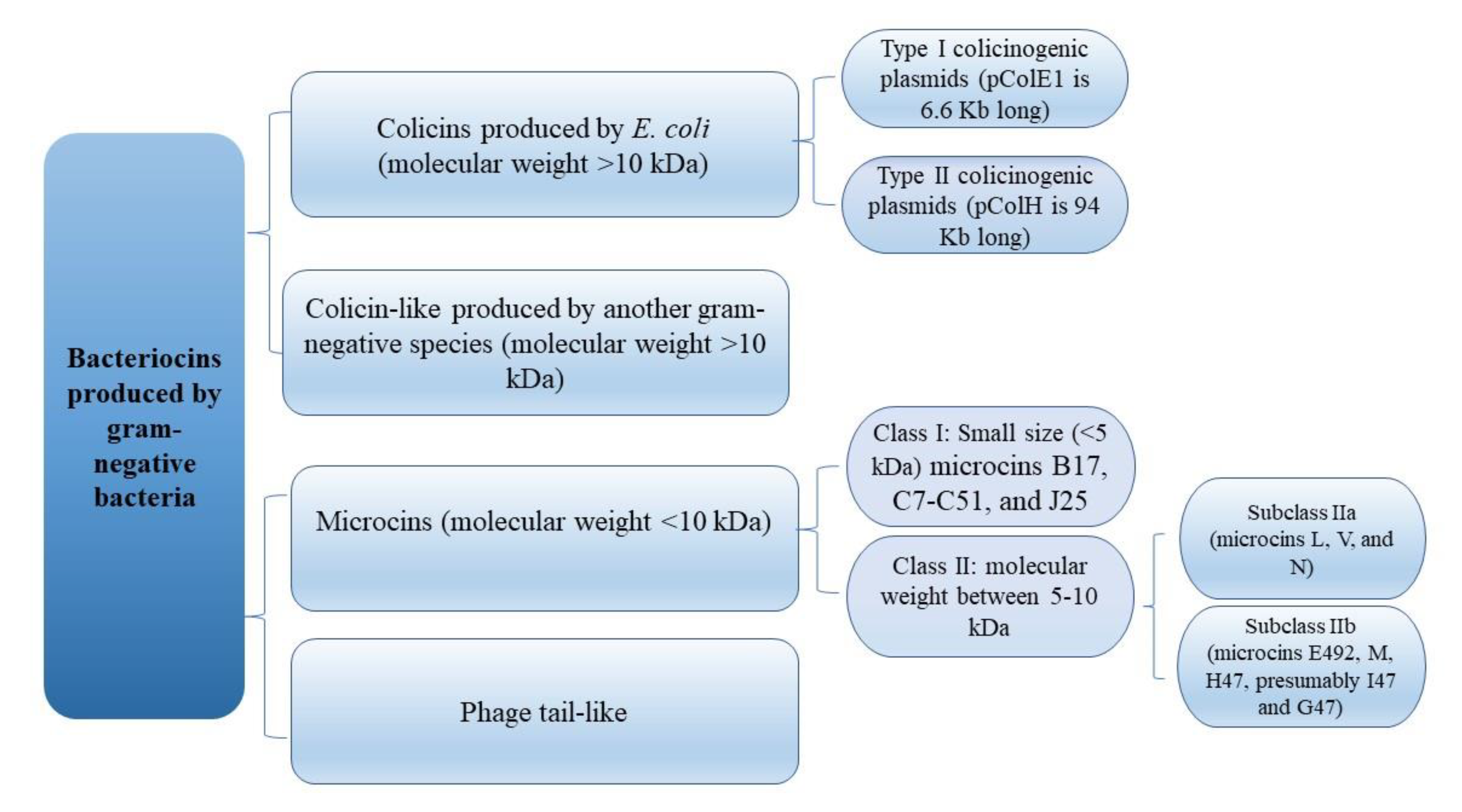
2. Colicins—Short Overview of Genetic Organization, Classification, and Mechanisms of Action
3. Microcins—Short Overview of Genetic Organization, Classification, and Mechanisms of Action
References
- Gillor, O.; Etzion, A.; Riley, M. The dual role of bacteriocins as anti-and probiotics. Appl. Microbiol. Biotechnol. 2008, 81, 591–606.
- Hegarty, J.W.; Guinane, C.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocin production: Arelatively unharnessed probiotic trait? F1000Research 2016, 5, 2587.
- Flaherty, R.A.; Freed, S.D.; Lee, S.W. The wide world of ribosomally encoded bacterial peptides. PLoS Pathog. 2014, 10, e1004221.
- Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Focus on modified microcins: Structural features and mechanisms of action. Biochimie 2002, 84, 511–519.
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubes, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229.
- Gordon, D.M.; O’Brien, C.L. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 2006, 152, 3239–3244.
- Paquette, S.J.; Zaheer, R.; Stanford, K.; Thomas, J.; Reuter, T. Competition among escherichia coli strains for space and resources. Veter. Sci. 2018, 5, 93.
- Cameron, A.; Zaheer, R.; Adator, E.H.; Barbieri, R.; Reuter, T.; McAllister, T.A. Bacteriocin occurrence and activity in Escherichia coli isolated from bovines and wastewater. Toxins 2019, 11, 475.
- Drider, D.; Rebuffat, S. Prokaryotic Antimicrobial Peptides: From Genes to Applications; Springer: New York, NY, USA, 2011.
- Konisky, J. Colicins and other bacteriocins with established modes of action. Annu. Rev. Microbiol. 1982, 36, 125–144.
- Braun, V.; Pilsl, H.; Gross, P. Colicins: Structures, modes of action, transfer through membranes and evolution. Arch. Microbiol. 1994, 161, 199–206.
- Duché, D.; Letellier, L.; Géli, V.; Bénédetti, H.; Baty, D. Quantification of group a colicin import sites. J. Bacteriol. 1995, 177, 4935–4939.
- Braun, V.; Patzer, S.I.; Hantke, K. Ton-dependent colicins and microcins: Modular Design and Evolution. Biochimie 2002, 84, 365–380.
- Šmarda, J.; Šmajs, D. Colicins-exocellular lethal proteins of Escherichia coli. Folia Microbiol. 1998, 43, 563–582.
- Cursino, L.; Smarda, J.; Chartone-Souza, E.; Nascimento, A.M.A. Recent updated aspects of colicins of Enterobacteriaceae. Braz. J. Microbiol. 2002, 33, 185–195.
- Hardy, K.G.; Meynell, G.G.; Dowman, J.E.; Spratt, B.G. Two Major Groups of Colicinogenic Factors: Their evolutionary significance. Mol. Gen. Genet. 1973, 125, 217–230.
- Riley, M.A. Positive selection for colicin diversity in bacteria. Mol. Biol. Evol. 1993, 10, 1048–1059.
- Herschman, H.R.; Helinski, D.R. Comparative study of the events associated with colicin induction. J. Bacteriol. 1967, 94, 691–699.
- Lu, F.M.; Chak, K.F. Two overlapping sos boxes in cole1 operon are responsible for the viability of cells harboring the col plasmid. Mol. Gen. Genet. 1996, 251, 407–411.
- Pugsley, A.P.; Schwartz, M.; Lavina, M.; Moreno, F. On the effect of ompr mutation on colicin e2 production. FEMS Microbiol. Lett. 1983, 19, 87–92.
- Ebina, Y.; Nakazawa, A. Cyclic AMP-dependent initiation and ρ-dependent termination of colicin e1 gene transcription. J. Biol. Chem. 1983, 258, 7072–7078.
- Dekker, N.; Tommassen, J.; Verheij, H.M. Bacteriocin release protein triggers dimerization of outer membrane phospholipase A In vivo. J. Bacteriol. 1999, 181, 3281–3283.
- Bénédetti, H.; Frenette, M.; Baty, D.; Knibiehler, M.; Pattus, F.; Lazdunski, J.C. Individual domains of colicins confer specificity in colicin uptake, in pore-properties and in immunity requirements. J. Mol. Biol. 1991, 217, 429–439.
- Davies, J.K.; Reeves, P. Genetics of resistance to colicins in Escherichia coli K12: Cross-resistance among resistance of group A. J. Bacteriol. 1975, 123, 102–117.
- Thomas, J.A.; Valvano, M.A. Role of tol genes in cloacin DF13 susceptibility of Escherichia Coli K-12 strains expressing the cloacin DF13- aerobactin receptor Lut A. J. Bacteriol. 1993, 175, 548–552.
- Ahmer, B.M.M.; Thomas, M.G.; Larsen, R.A.; Postle, K. Characterization of the exbBD operon of Escherichia coli and role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 1995, 177, 4742–4747.
- Michel-Briand, Y.; Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510.
- Baquero, F.; Lanza, V.F.; Baquero, M.; Campo, R.; Bravo-Vázquez, A.D. Microcins in Enterobacteriaceae: Peptide antimicrobials in the eco-active intestinal chemosphere. Front. Microbiol. 2019, 10, 2261.
- Barreteau, H.; Bouhss, A.; Gérard, F.; Duché, D.; Boussaid, B.; Blanot, D.; Lloubes, R.; Mengin-Lecreulx, D.; Touzé, T. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II degrading enzyme. J. Biol. Chem. 2010, 285, 12378–12389.
- Cavard, D.; Sauve, P.; Heitz, F.; Pattus, F.; Martinez, C.; Dijkman, R.; Lazdunski, C. Hydrodynamic properties of colicin A. existence of a high-affinity lipid-binding site and oligomerization at acid pH. Eur. J. Biochem. 1988, 172, 507–512.
- Schein, S.J.; Kagan, B.L.; Finkelstein, A. Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature 1978, 276, 159–163.
- Parker, M.W.; Pattus, F.; Tucker, A.D.; Tsernoglou, D. Structure of the membrane pore-forming fragments of colicin A. Nature 1989, 337, 93–96.
- Lakey, J.H.; Duche, D.; Gonzalez-Manas, J.M.; Baty, D.; Pattus, F. Fluorescence energy transfer distance measurements: The hydrophobic helical hairpin of colicin a in the membrane bound state. J. Mol. Biol. 1993, 230, 1055–1067.
- Slatin, S.L.; Duche, D.; Kienker, P.K.; Baty, D. Gating movements of colicin A and Colicin Ia are different. J. Membr. Biol. 2004, 202, 73–83.
- Collarini, M.; Amblard, G.; Lazdunski, C.; Pattus, F. Gating processes of channels induced by colicin A, its C-terminal fragment and colicin E1, in planar lipid bilayers. Eur. Biophys. J. 1987, 14, 147–153.
- Nogueira, R.A.; Varanda, W.A. Gating properties of channels formed by colicin Ia in planar lipid bilayer membranes. J. Membr. Biol. 1988, 105, 143–153.
- James, R.; Penfold, C.N.; Moore, G.R.; Kleanthous, C. Killing of E. coli cells by E group nuclease colicins. Biochimie 2002, 84, 381–389.
- Ku, W.Y.; Liu, Y.W.; Hsu, Y.C.; Liao, C.C.; Liang, P.H.; Yuan, H.S.; Chak, K.F. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 2002, 30, 1670–1678.
- Krone, W.J.A.; de Vries, P.; Koningstein, G.; de Jonge, A.J.R.; de Graaf, F.K.; Oudega, B. uptake of cloacin DF13 by susceptible cells: Removal of immunity protein and fragmentation of cloacin molecules. J. Bacteriol. 1986, 166, 260–268.
- Chérier, D.; Patin, D.; Blanot, D.; Touzé, T.; Barreteau, H. The biology of colicin m and its orhologs. Antibiotics 2021, 10, 1109.
- Braun, V.; Schaller, K.; Wabl, M.R. Isolation, characterization, and action of colicin M. Antimicrob. Agents Chemother. 1974, 5, 520–533.
- Harkness, R.E.; Braun, V. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J. Biol. Chem. 1989, 264, 6177–6182.
- Harkness, R.E.; Olschläger, T. The biology of colicin M. FEMS Microbiol. Rev. 1991, 8, 27–41.
- Baquero, F.; Moreno, F. The microcins. FEMS Microbiol. Lett. 1984, 23, 117–124.
- Duquesne, S.; Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 2007, 24, 708–734.
- Duquesne, S.; Petit, V.; Peduzzi, J.; Rebuffat, S. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J. Mol. Microbiol. Biotechnol. 2007, 13, 200–209.
- Severinov, K.; Semenova, E.; Kazakov, A.; Kazakov, T.; Gelfand, M.S. Low-molecular-weight post-translationally modified microcins. Mol. Microbiol. 2007, 65, 1380–1394.
- Poey, M.E.; Azpiroz, M.F.; Laviña, M. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 2006, 50, 1411–1418.
- Vassiliadis, G.; Destoumieux-Garzón, D.; Lombard, C.; Rebuffat, S.; Peduzzi, J. Siderophore microcins form the first family of structure-related antimicrobial peptides from Enterobacteriaceae:isolation and characterization of microcins M and H47. Antimicrob. Agents Chemother. 2010, 54, 288–297.
- Rebuffat, S. Microcins in action: Amazing defence strategies of Enterobacteria. Biochem. Soc. Trans 2012, 40, 1456–1462.
- Lagos, R.; Villanueva, J.E.; Monasterio, O. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 1999, 181, 212–217.
- Lagos, R.; Baeza, M.; Corsini, G.; Hetz, C.; Strahsburger, E.; Castillo, J.A.; Vergara, C.; Monasterio, O. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 2011, 42, 229–243.
- Thomas, X.; Destoumieux-Garzon, D.; Peduzzi, J.; Afonso, C.; Blond, A.; Biriirakis, N.; Goulard, C.; Dubost, L.; Thai, R.; Tabet, J.C.; et al. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 2004, 279, 28233–28242.
- Rebuffat, S. Microcins. In Handbook of Biologically Active Peptides, 2nd ed.; Kastin, A.J., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 129–137.
- Pons, A.M.; Delalande, F.; Duarte, M.; Benoit, S.; Lanneluc, I.; Sablé, S.; Van Dorsselaer, A.; Cottenceau, G. Genetic analysis and complete primary structure of microcin L. Antimicrob. Agents Chemother. 2004, 48, 505–513.
- Fomenko, E.D.; Metlitskaya, A.Z.; Péduzzi, J.; Goulard, C.; Katrukha, G.S.; Gening, L.V.; Rebuffat, S.; Khmel, I.A. Microcin C51 plasmid genes: Possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 2003, 47, 2868–2874.
- Allali, N.; Afif, H.; Couturier, M.; Van Melderen, L. The highly conserved TldD and TldE proteins of Escherichia coli are involved in microcin B17 processing and in CcdA degradation. J. Bacteriol. 2002, 12, 3224–3231.
- Morin, N.; Lanneluc, I.; Connil, N.; Cottenceau, M.; Pons, A.M.; Sablé, S. Mechanism of bactericidal activity of microcin L in Escherichia coli and Salmonella enterica. Antimicrob. Agents Chemother. 2011, 55, 997–1007.
- Rebuffat, S. Microcins from Enterobacteria: On the edge between Gram-positive bacteriocins and colicins. In Prokaryotic Antimicrobial Peptides: From Genes to Applications; Drider, D., Rebuffat, S., Eds.; Springer: New York, NY, USA, 2011; pp. 333–353.
- Pierrat, O.A.; Maxwell, A. Evidence for the role of DNA strand passage in the mechanism of action of microcin B17 on DNA gyrase. Biochemistry 2005, 44, 4204–4215.
- Mukhopadhyay, J.; Sineva, E.; Knight, J.; Levy, R.M.; Ebright, R.H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 2004, 14, 739–751.
- Rodríguez, E.; Laviña, M. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 2003, 47, 181–187.
- Scholl, D. Phage Tail-Like Bacteriocins. Annu. Rev. Virol. 2017, 4, 453–467.
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639.




