Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zhihao Chen | -- | 2420 | 2022-10-10 09:17:36 | | | |
| 2 | Camila Xu | Meta information modification | 2420 | 2022-10-10 09:53:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chen, Z.; Huai, Y.; Mao, W.; Wang, X.; Ru, K.; Qian, A.; Yang, H. Liquid–Liquid Phase Separation in Metabolic Diseases. Encyclopedia. Available online: https://encyclopedia.pub/entry/28675 (accessed on 08 March 2026).
Chen Z, Huai Y, Mao W, Wang X, Ru K, Qian A, et al. Liquid–Liquid Phase Separation in Metabolic Diseases. Encyclopedia. Available at: https://encyclopedia.pub/entry/28675. Accessed March 08, 2026.
Chen, Zhihao, Ying Huai, Wenjing Mao, Xuehao Wang, Kang Ru, Airong Qian, Hong Yang. "Liquid–Liquid Phase Separation in Metabolic Diseases" Encyclopedia, https://encyclopedia.pub/entry/28675 (accessed March 08, 2026).
Chen, Z., Huai, Y., Mao, W., Wang, X., Ru, K., Qian, A., & Yang, H. (2022, October 10). Liquid–Liquid Phase Separation in Metabolic Diseases. In Encyclopedia. https://encyclopedia.pub/entry/28675
Chen, Zhihao, et al. "Liquid–Liquid Phase Separation in Metabolic Diseases." Encyclopedia. Web. 10 October, 2022.
Copy Citation
Liquid–liquid phase separation (LLPS) is a reversible and dynamical biophysical process where homogeneous biomacromolecules spontaneously de-mix into two coexisting liquid phases (a condensed phase and a dilute phase) through transient multivalent macromolecular interactions.
Alzheimer’s disease (AD)
liquid–liquid phase separation (LLPS)
membraneless organelles (MLOs)
metabolic bone diseases (MBDs)
1. Introduction
Liquid–liquid phase separation (LLPS) is a reversible and dynamical biophysical process where homogeneous biomacromolecules spontaneously de-mix into two coexisting liquid phases (a condensed phase and a dilute phase) through transient multivalent macromolecular interactions [1][2]. Currently, LLPS is reported to be considered the underlying of multiple biological processes, especially for the formation of membraneless organelles (MLOs), such as processing bodies (P-bodies), stress granules, and nucleolar. In fact, LLPS tends to compartmentalize and concentrate biomacromolecules into liquid-like condensates, which underlies MLO formation to explain the self-assembly process of subcellular structures [3]. LLPS also serves as an important natural defense mechanism in response to external various stimuli in living cells [4]. It is reported that LLPS is associated with the pathogenesis of multiple human diseases, such as neurodegeneration, infectious diseases, cancer, and aging diseases [5][6].
Metabolic diseases usually disrupt the critical biochemical reactions of cells, including the processing or transport of proteins (amino acids), carbohydrates (sugars and starches), or lipids (fatty acids) [7]. Most importantly, several metabolic diseases are strongly associated with amyloid depositions which are insoluble proteinaceous aggregates depositions.
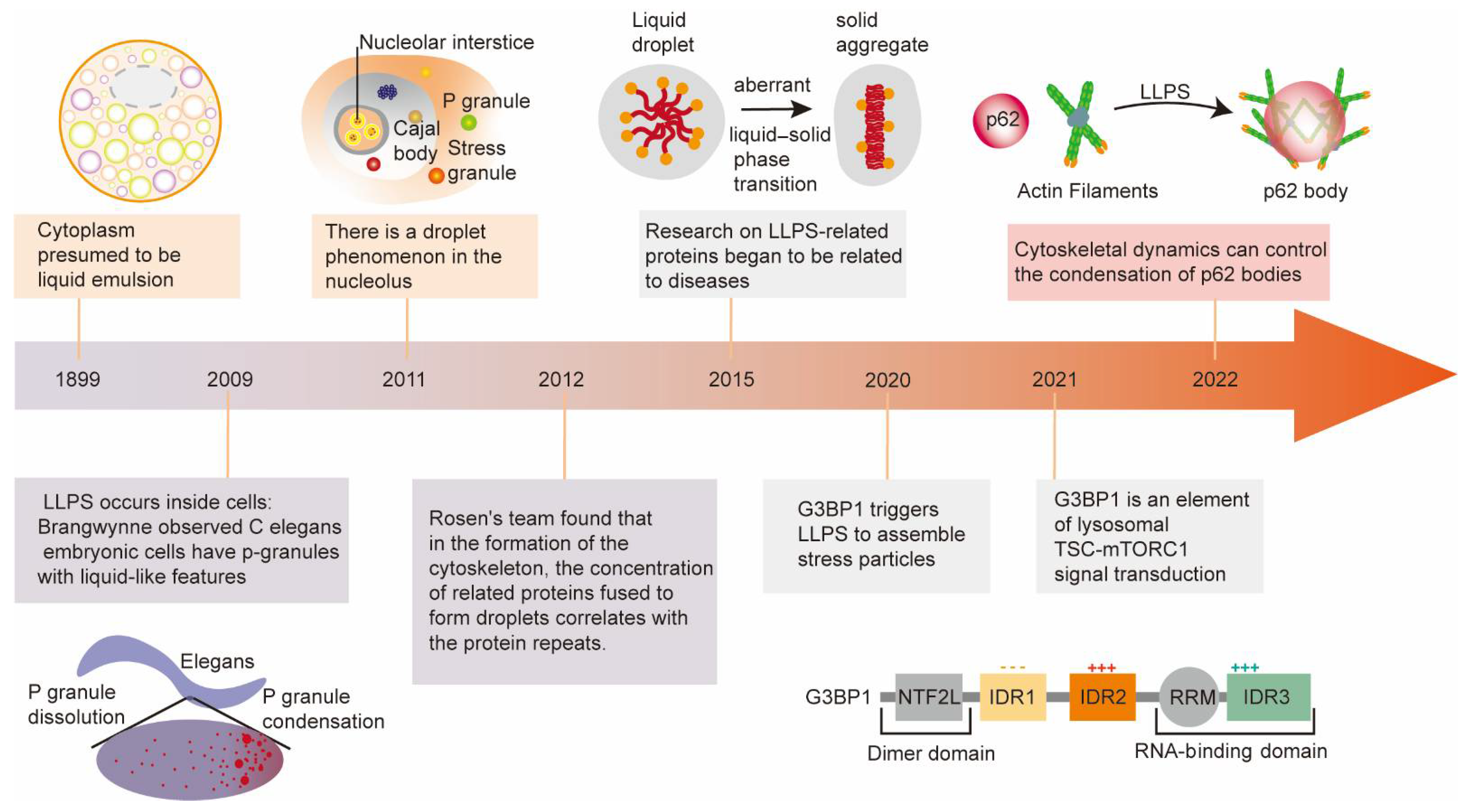

2. Major Milestones of Liquid–Liquid Phase Separation Development
Although increasing research on LLPS has identified its essential roles in physiology and diseases, it also experienced a tortuous development history. The following brief introduction will discuss the research development and milestone achievements of liquid–liquid phase separation (Figure 1).

Figure 1. The development history and discovery of the amazing and vital roles of LLPS in biology. Representative milestones sparking tremendous development of LLPS are enumerated in the figure.
3. Representative Research Methods of Liquid–Liquid Phase Separation
At present, LLPS has become a research hotspot in the field of biology. Therefore, the research methods of LLPS are also gradually diversified. Here, researchers briefly introduce the existing representative strategies of LLPS based on the in vitro and in vivo reported research, respectively.
3.1. In Vitro
It is easy to observe the process of LLPS and control the concentration and environmental conditions of each component in vitro. Thus, diverse microscopies are increasingly applied to determine the characteristics of liquid-like droplets formed by LLPS. For example, differential interference contrast (DIC) imaging is a representative method to visualize the properties of droplets, which can present the coexistence of two or more phases. Besides, fluorescence correlation spectroscopy (FCS) is considered another ideal tool to estimate the diffusion capacity of a single molecule inside the LLPS droplet [8]. FCS is always used to detect sparsely labeled and highly mobile components as well as droplet dilution. Atomic force microscopy (AFM) can describe the properties of biological condensate materials, such as viscosity, pore size, elasticity, and other parameters. Zeng et al. measured the mechanical properties of postsynaptic density (PSD) droplets to monitor individual phase performance by AFM [9]. Furthermore, liquid-phase transmission electron microscopy (LP-TEM) can enable direct visualization and real-time observation of liquid-like droplets formation to discover and renew biological assembly mechanisms [10]. Moreover, the fluorescent labeling and the dynamic imaging of liquid-like droplets are powerful methods to study the mechanisms of LLPS formation. In addition, the turbidity measurement assay is also a popular intuitive detection method for LLPS in vitro [11][12]. The components in the solution can scatter visible light from tens to hundreds of nanometers in diameter, which could be measured by optical density. Notably, this method just only detects the components in a droplet, the observation of the droplet shape, size and formation principle still requires a combination with microscopy [13]. Furthermore, centrifugal precipitation is also another common detection strategy of LLPS. It can be observed transparent droplets that differ from the sediment and assess the proteins in different phases through centrifugation precipitation [14]. The light phase and the dense phase were separated by centrifugation, and then their concentration is measured by spectroscopy. Fluorescence recovery after photobleaching (FRAP) allows to capture of the exchange of substances between dense and dilute phases and observe the constant dynamic change process of LLPS, which is increasingly used to demonstrate molecular motion inside droplets [15]. Additionally, the optoDroplet is a tool that uses light to manipulate matter inside living cells and has begun to explain how proteins assemble into different liquid and gel-like solid states, a key to understanding many critical cellular operations. The optoDroplet tool is starting to allow to dissect the rules of physics and chemistry that govern the self-assembly of MLOs. Importantly, researchers have only introduced the most common research methods of LLPS and MLOs. There are many more approaches to examine LLPS and MLOs, such as passive microrheology, active microrheology, cryoelectron tomography, nuclear magnetic resonance, capillary flow experiments, microfluidic tools as well as Corelets and PixELL platforms [16]. The existing research methods of LLPS in vitro are diversified (Figure 2), and more accurate detection technologies still need to be developed in the future.

Figure 2. Representative research methods and technology to identify or study LLPS. Various microscopic techniques can be used to detect the process of phase transition and visualize the properties of droplets. Centrifugal precipitation is another common detection strategy of LLPS. The FRAP is the well-recognized method for the observation of LLPS, which was accomplished by measuring the fluorescence intensity of the bleached region prior to, immediately after, and throughout recovery from bleaching. The optoDroplet provides a level of control that can be used to precisely map the phase diagram in living cells.
3.2. In Vivo
The research methods of LLPS in vivo are more complicated compared with that in vitro. The high protein concentration is one of the important prerequisites for LLPS in cells. Therefore, overexpression of LLPS-triggering proteins is the common manner to drive and detect LLPS in vivo. At present, it is believed that the accepted criteria for a phase separation structure are the formation of a spherical structure, the ability to fuse, and the ability to recover from photobleaching [13]. The recovery time of FRAP not only depends on the protein/RNA concentration but also on the droplet size and the bleaching area [13]. Therefore, combining it with the other methods is more accurate for LLPS detection (Figure 2).
To identify the property of LLPS in vivo, Delarue et al. developed a homomultimeric scaffold fused with a fluorescent protein, named genetically encoded nanoparticles (GEMS), and used as an effective probe in the cytoplasmic matrix. The probe can evaluate the condensate porosity and parameters in the cellular environment [17]. Compared with the numerous research strategies of LLPS in vitro, how determining the physical and chemical properties of phase separation droplets in vivo still needs further exploration. In addition, it is also needful to explore several novel methods to explore the biological functions of LLPS in cells.
4. Liquid–Liquid Phase Separation Underlies MLOs Formation
It is well recognized that LLPS of biomacromolecules have emerged as a biophysical basis for the formation of MLOs in living cells [15]. Ubiquitously, MLOs in eukaryotic cells modulate a variety of physiological and pathological traits through multiple ways, which are closely related to the physical properties, types, and intracellular localization of MLOs [18]. Moreover, MLOs formed by LLPS are broadly distributed in the cytoplasm, nucleus, and membrane [19][20]. In this section, researchers mainly review the biological function of the MLOs localized in the cytoplasm such as stress granules (SGs), processing bodies (P-bodies), as well as in the nucleus including nucleoli, paraspeckles, PML bodies, and Cajal bodies. Table 1 displayed representative MLOs with different cellular localization and their function.
Table 1. Examples of the various MLOs formed by LLPS and their functions.
| Localization | Names of Condensates | Biological Function | References |
|---|---|---|---|
| Plasma membrane | TCR clusters | T-cell immune signal transduction | [21] |
| Nephrin clusters | Glomerular filtration barrier | [22] | |
| Actin patches | Endocytosis | [23] | |
| Focal adhesions | Cell adhesion and migration | [24] | |
| Synaptic densities | Neurotransmission | [14] | |
| Cytoplasm | Stress granules | mRNA storage and translational regulation | [25] |
| RNA transport granules | mRNA storage and transport in neuronal cells | [26] | |
| U body | Storage and assembly of snRNPs | [27] | |
| P body | mRNA decay and silencing | [28] | |
| Balbiani body | A transient collection of proteins, RNA, and membrane-bound organelles found in primary oocytes of all animals observed to date | [29] | |
| P granules | Germ cell lineage maintenance in Caenorhabditis elegans | [15] | |
| Nucleus | cGAS condensates | Innate immune signaling | [30] |
| Cleavage body | mRNA processing | [31] | |
| Cajal body | Assembling spliceosomal small nuclear ribonucleoproteins | [31][32] | |
| Nucleoli | rRNA storage, rRNA synthesis and processing, and assembly of ribosomal subunits | [32] | |
| Gem | Aid histone mRNA processing | [33] | |
| Nuclear speckles | mRNA splicing | [34] | |
| OPT domain | Transcriptional regulation | [35] | |
| PcG body | Transcriptional repression | [36] | |
| PML bodies | Apoptotic signaling, anti-viral defense, and transcription regulation | [37] | |
| Histone locus body | Processing of histone mRNAs | [38] | |
| Paraspeckles | Storage of certain RNAs | [39] | |
| Perinucleolar compartment | Related to malignancy | [40] |
4.1. Cytoplasmic-Localized MLOs
Cytoplasmic-localized MLOs are dynamically assembled by the LLPS driving the temporarily untranslated RNAs and proteins, which coalesce into a concentrated state (the condensed phase) in the cytoplasm. Prominent examples of cytoplasmic-localized MLOs mainly include the stress granules (SGs), the processing bodies (P-bodies), the RNA transport granules, and the germ granules.
The stress granules (SGs) are a predominant type of cytoplasmic-localized MLOs formed by the crowded protein and RNA. The SGs immediately start to accumulate and regulate the mRNA utilization in eukaryotic cells under stress, which is essential for maintaining cell integrity and intracellular homeostasis [25][41]. Additionally, SG components mainly include aggregation-prone RNA binding proteins (RBPs), protein kinases, RNA helicases, structural constituents of ribosomes, calcium-binding proteins, hydrolases, and cytoskeletal proteins [42][43]. Moreover, dynein, microtubules [44], and various nucleocytoplasmic shuttling RBPs (TIA-1, TIAR, and HUR) [42] assist in the SGs secondary aggregation and assembly, which determines the speed and size of SG assembly. Moreover, the SGs are highly dynamic in nature, assembling, and dissembling quickly upon stress induction or stress disappearance, respectively. Their dynamic properties are mainly highlighted by the cytoskeleton system, which is a scaffold for SGs’ dynamic maintenance and movement [45]. Numerous researchers found that maintaining a proper SG dynamic might be a potential strategy to ensure cellular homeostasis and normal biological function in the living cell [46]. Thus, the normal dynamics of SGs play an important role in responding to stress stimuli, which can otherwise induce various human diseases.
The processing bodies (P-bodies) are the highly conserved cytoplasmic foci with properties of liquid droplets, which are formed by LLPS and are primarily composed of translation-arrested RNAs and RBPs related to mRNA decay [28][47]. Moreover, the P-bodies purification revealed that multiple RBPs including HNRNPU, IGF2BP1, DHX9, and HNRNPQ were the core components of P-bodies [48][49]. Additionally, P-bodies are distinct from SGs in multiple aspects, including the formation conditions, morphology, function, as well as components. Specifically, unlike SGs being exclusively stress-induced, the P-bodies are constitutive in some cells and nevertheless increase in size and number in response to stress [50]. Therefore, researchers conclude from these studies that the P-bodies exert multiple regulatory roles in the post-transcriptional processes, translation repression, and mRNA decay machinery.
In addition to SGs and P-bodies, there are still other well-studied cytoplasmic-localized MLOs, including germ granules, RNA transport granules, P granules, and the Balbiani body. For example, the germ granules and the P granules are conserved condensates enriched for RNA and RBPs in the germ cell cytoplasm, which play essential roles in the mRNA translation during gametogenesis and embryonic development [51]. The Balbiani body, also called a membraneless ball of mitochondria, contains various biomacromolecules and numerous membranous organelles. The Balbiani body is widely present in the majority of mammal oocytes [52]. In summary, these findings emphasize the important roles of LLPS in the formation and maintenance of cytoplasmic-localized MLOs and confirm the biological function of MLOs in cell growth and development.
4.2. Nuclear-Localized MLOs
In addition to the multiple cytoplasmic-localized MLOs, LLPS also is important for driving the assembly of various nuclear-localized MLOs such as nucleoli, Cajal bodies, and nuclear speckles, and underlies their biogenesis. The condensates within the nucleus could directly interact with chromatin, and thus potentially control its organization and gene expression. Moreover, the biomacromolecules and their multiple unique domains help to build these nuclear-localized condensates in the nucleus. In the following section, researchers will detail the assembly of the proteins/RNA in multiple nuclear-localized condensates and discuss the biological functions of the nuclear-localized MLOs.
The nucleolus is the most prototypical and prominent nuclear MLO. Nucleolus forms around the chromosome regions containing stretches of tandem ribosomal DNA (rDNA) gene repeats, known as nucleolar organizer regions (NORs) [53]. There is evidence that the nucleolus is formed through LLPS by its macromolecular components and exerts dynamic and liquid-like physical properties which might facilitate functions of the nucleolus in ribosome biogenesis and cellular stress sense [54]. Indeed, the nucleolus is composed of various RNA and hundreds of different proteins including RBPs. Nucleolin, a multifunctional stress-responsive RBP, is abundant in the nucleolus. It is reported that nucleolin could participate in rDNA transcription, rRNA maturation, ribosome assembly, and nucleocytoplasmic transport. Furthermore, nucleolin contains four RNA binding motifs, which indicates that nucleolin could undergo LLPS through its multivalent interactions with many other RNAs, and thus mediate the assembly of the nucleolus [55]. Besides, another nuclear protein nucleophosmin (NPM) has been confirmed to be able to facilitate the LLPS process in the multilayered structure of the nucleolus [56]. Furthermore, NPM harbors a low-complexity domain bound by poly (GR) and poly (PR), which could alter the LLPS properties of NPM and thus influence nucleolar dynamics in cells [57]. In brief, the LLPS triggered by several nuclear proteins plays crucial roles in the formation and biological function of the nucleolus.
Nuclear speckles, another well-studied MLO formed by LLPS in nuclear, exhibit dynamic and irregular shapes. Nuclear speckles are subnuclear structures enriched in the RBPs involved in splicing, which are located in the interchromatin regions of the nucleoplasm in mammalian cells [58]. Furthermore, nuclear speckles are formed through the exchange of constituent RBPs and RNAs with the surrounding nucleoplasm [34]. In addition, the nuclear speckles are reported to be enriched for the SP protein family, which is a set of RBPs named for the IDRs of their serine and arginine residues. For instance, SRRM2, an important RBP in the SR family, was found as a core nuclear speckle scaffold protein, which is required for nuclear speckle formation [59]. In summary, nuclear speckles are one type of self-assembled MLO composed of LLPS-related RBPs or RNAs that mediate multiple critical steps of RNA processing.
Taken together, these findings have highlighted the important roles of LLPS in the formation of MLOs in the nucleus, as well as the LLPS of biomacromolecules participants in the heterochromatin formation and coordination of mRNA processing in the eukaryotic nucleus. Despite LLPS being involved in the physiological formation and maintenance of various MLOs, LLPS triggered by abnormal or mutated proteins is also linked to pathology due to irreversible hydrogelation through amyloid-like aggregation.
References
- Su, Q.; Mehta, S.; Zhang, J. Liquid-liquid phase separation: Orchestrating cell signaling through time and space. Mol. Cell 2021, 81, 4137–4146.
- Tong, X.; Tang, R.; Xu, J.; Wang, W.; Zhao, Y.; Yu, X.; Shi, S. Liquid-liquid phase separation in tumor biology. Signal Transduct. Target. Ther. 2022, 7, 221.
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298.
- Tsang, B.; Pritisanac, I.; Scherer, S.W.; Moses, A.M.; Forman-Kay, J.D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 2020, 183, 1742–1756.
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid–liquid phase separation in human health and diseases. Signal Transduct. Target. Ther. 2021, 6, 290.
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213.
- Hoffman, D.J.; Powell, T.L.; Barrett, E.S.; Hardy, D.B. Developmental origins of metabolic diseases. Physiol. Rev. 2021, 101, 739–795.
- Wei, M.-T.; Elbaum-Garfinkle, S.; Holehouse, A.S.; Chen, C.C.-H.; Feric, M.; Arnold, C.B.; Priestley, R.D.; Pappu, R.V.; Brangwynne, C.P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017, 9, 1118–1125.
- Zeng, M.; Chen, X.; Guan, D.; Xu, J.; Wu, H.; Tong, P.; Zhang, M. Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell 2018, 174, 1172–1187.e1116.
- Rizvi, A.; Mulvey, J.T.; Patterson, J.P. Observation of liquid-liquid-phase separation and vesicle spreading during supported bilayer formation via liquid-phase transmission electron microscopy. Nano Lett. 2021, 21, 10325–10332.
- Milovanovic, D.; Wu, Y.; Bian, X.; De Camilli, P. A liquid phase of synapsin and lipid vesicles. Science 2018, 361, 604–607.
- Wang, H.; Yan, X.; Aigner, H.; Bracher, A.; Nguyen, N.D.; Hee, W.Y.; Long, B.M.; Price, G.D.; Hartl, F.U.; Hayer-Hartl, M. Rubisco condensate formation by ccmm in β-carboxysome biogenesis. Nature 2019, 566, 131–135.
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419–434.
- Zeng, M.L.; Shang, Y.; Araki, Y.; Guo, T.F.; Huganir, R.L.; Zhang, M.J. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 2016, 166, 1163–1175.
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline p granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732.
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid–liquid phase separation as an organizing principle of intracellular space: Overview of the evolution of the cell compartmentalization concept. Cell. Mol. Life Sci. 2022, 79, 251.
- Delarue, M.; Brittingham, G.P.; Pfeffer, S.; Surovtsev, I.; Pinglay, S.; Kennedy, K.; Schaffer, M.; Gutierrez, J.; Sang, D.; Poterewicz, G. Mtorc1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 2018, 174, 338–349.e320.
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127.
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional control of phase-separated cellular bodies. Cell 2016, 166, 651–663.
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881.
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase separation of signaling molecules promotes t cell receptor signal transduction. Science 2016, 352, 595–599.
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340.
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and endocytosis in budding yeast. Genetics 2015, 199, 315–358.
- Shan, Z.L.; Tu, Y.T.; Yang, Y.; Liu, Z.H.; Zeng, M.L.; Xu, H.S.; Long, J.F.; Zhang, M.J.; Cai, Y.; Wen, W.Y. Basal condensation of numb and pon complex via phase transition during drosophila neuroblast asymmetric division. Nat. Commun. 2018, 9, 737.
- Yang, P.G.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.P.; Messing, J.; Yurtsever, U.; Yang, Z.M.; Wu, J.J.; Li, Y.X.; Pan, Q.F.; et al. G3bp1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 2020, 181, 325–345.e328.
- Hofweber, M.; Hutten, S.; Bourgeois, B.; Spreitzer, E.; Niedner-Boblenz, A.; Schifferer, M.; Ruepp, M.D.; Simons, M.; Niessing, D.; Madl, T.; et al. Phase separation of fus is suppressed by its nuclear import receptor and arginine methylation. Cell 2018, 173, 706–719.e713.
- Liu, J.L.; Gall, J.G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with p bodies. Proc. Natl. Acad. Sci. USA 2007, 104, 11655–11659.
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mrna degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286.
- Nojima, H.; Rothhamel, S.; Shimizu, T.; Kim, C.H.; Yonemura, S.; Marlow, F.L.; Hibi, M. Syntabulin, a motor protein linker, controls dorsal determination. Development 2010, 137, 923–933.
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cgas activates innate immune signaling. Science 2018, 361, 704–709.
- Li, L.; Roy, K.; Katyal, S.; Sun, X.J.; Bleoo, S.; Godbout, R. Dynamic nature of cleavage bodies and their spatial relationship to ddx1 bodies, cajal bodies, and gems. Mol. Biol. Cell 2006, 17, 1126–1140.
- Handwerger, K.E.; Cordero, J.A.; Gall, J.G. Cajal bodies, nucleoli, and speckles in the xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 2005, 16, 202–211.
- Cauchi, R.J. Gem formation upon constitutive gemin3 overexpression in drosophila. Cell Biol. Int. 2011, 35, 1233–1238.
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646.
- Harrigan, J.A.; Belotserkovskaya, R.; Coates, J.; Dimitrova, D.S.; Polo, S.E.; Bradshaw, C.R.; Fraser, P.; Jackson, S.P. Replication stress induces 53bp1-containing opt domains in g1 cells. J. Cell Biol. 2011, 193, 97–108.
- Pirrotta, V.; Li, H.B. A view of nuclear polycomb bodies. Curr. Opin. Genet. Dev. 2012, 22, 101–109.
- Lallemand-Breitenbach, V.; de The, H. Pml nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661.
- Nizami, Z.; Deryusheva, S.; Gall, J.G. The cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010, 2, a000653.
- Hennig, S.; Kong, G.; Mannen, T.; Sadowska, A.; Kobelke, S.; Blythe, A.; Knote, G.J.; Iyer, K.S.; Ho, D.W.; Newcombe, E.A.; et al. Prion-like domains in rna binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015, 210, 529–539.
- Pollock, C.; Huang, S. The perinucleolar compartment. Cold Spring Harb. Perspect. Biol. 2010, 2, a000679.
- Kedersha, N.; Anderson, P. Stress granules: Sites of mrna triage that regulate mrna stability and translatability. Biochem. Soc. Trans. 2002, 30, 963–969.
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nobrega, C. Stress granules, rna-binding proteins and polyglutamine diseases: Too much aggregation? Cell Death Dis. 2021, 12, 592.
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The integral role of rna in stress granule formation and function. Front. Cell Dev. Biol. 2021, 9, 621779.
- Bartoli, K.M.; Bishop, D.L.; Saunders, W.S. The role of molecular microtubule motors and the microtubule cytoskeleton in stress granule dynamics. Int. J. Cell Biol. 2011, 2011, 939848.
- Nadezhdina, E.S.; Lomakin, A.J.; Shpilman, A.A.; Chudinova, E.M.; Ivanov, P.A. Microtubules govern stress granule mobility and dynamics. BBA-Mol. Cell Res. 2010, 1803, 361–371.
- Cao, X.L.; Jin, X.J.; Liu, B.D. The involvement of stress granules in aging and aging-associated diseases. Aging Cell 2020, 19, e13136.
- Luo, Y.; Na, Z.K.; Slavoff, S.A. P-bodies: Composition, properties, and functions. Biochemistry 2018, 57, 2424–2431.
- Hubstenberger, A.; Courel, M.; Benard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-body purification reveals the condensation of repressed mrna regulons. Mol. Cell 2017, 68, 144–157.
- Standart, N.; Weil, D. P-bodies: Cytosolic droplets for coordinated mrna storage. Trends Genet. 2018, 34, 612–626.
- Riggs, C.L.; Kedersha, N.; Ivanov, P.; Anderson, P. Mammalian stress granules and p bodies at a glance. J. Cell Sci. 2020, 133, jcs242487.
- Sengupta, M.S.; Boag, P.R. Germ granules and the control of mrna translation. IUBMB Life 2012, 64, 586–594.
- Kloc, M.; Jedrzejowska, I.; Tworzydlo, W.; Bilinski, S.M. Balbiani body, nuage and sponge bodies—the germ plasm pathway players. Arthropod. Struct. Dev. 2014, 43, 341–348.
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying r-rich linear motifs and rrna. Elife 2016, 5, e13571.
- Feric, M.; Vaidya, N.; Harmon, T.; Mitrea, D.; Zhu, L.; Richardson, T.; Kriwacki, R.; Pappu, R.; Brangwynne, C. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686–1697.
- Jia, W.Y.; Yao, Z.Y.; Zhao, J.J.; Guan, Q.B.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (ncl). Life Sci. 2017, 186, 1–10.
- Mitrea, D.M.; Cika, J.A.; Stanley, C.B.; Nourse, A.; Onuchic, P.L.; Banerjee, P.R.; Phillips, A.H.; Park, C.G.; Deniz, A.A.; Kriwacki, R.W. Self-interaction of npm1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 2018, 9, 842.
- Lee, K.H.; Zhang, P.P.; Kim, H.J.; Mitrea, D.M.; Sarkar, M.; Freibaum, B.D.; Cika, J.; Coughlin, M.; Messing, J.; Molliex, A.; et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 2016, 167, 774–788.
- Ilik, I.A.; Aktas, T. Nuclear speckles: Dynamic hubs of gene expression regulation. FEBS J. 2021. early view.
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome—phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093.
More
Information
Subjects:
Endocrinology & Metabolism
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
10 Oct 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





