| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jose De La Fuente | + 2263 word(s) | 2263 | 2020-02-07 09:56:43 | | | |
| 2 | Jose De La Fuente | + 2411 word(s) | 2411 | 2020-02-07 10:42:25 | | | | |
| 3 | Jose De La Fuente | + 13 word(s) | 2424 | 2020-02-09 09:19:40 | | | | |
| 4 | Dean Liu | -703 word(s) | 1721 | 2020-10-29 10:41:43 | | | | |
| 5 | Dean Liu | -703 word(s) | 1721 | 2020-10-29 10:42:43 | | |
Video Upload Options
The main objective of this study was to propose a novel methodology to approach challenges in molecular biology. Akirin/Subolesin (AKR/SUB) are vaccine protective antigens and a model for the study of the interactome due to its conserved function in the regulation of different biological processes such as immunity and development throughout the metazoan. Herein, three visual artists and a music professor collaborated with scientists for the functional characterization of the AKR2 interactome in the regulation of the NF-kB pathway in human placenta cells. The results served as a methodological proof-of-concept to advance this research area. The results showed new perspectives on unexplored characteristics of AKR2 with functional implications. These results included protein dimerization, the physical interactions with different proteins simultaneously to regulate various biological processes defined by cell type-specific AKR-protein interactions and how these interactions positively or negatively regulate the Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-kB) signaling pathway in a biological context-dependent manner. These results suggested that AKR2 interacting proteins might constitute suitable secondary transcription factors for cell and stimulus-specific regulation of NF-kB. Musical perspective supported AKR/SUB evolutionary conservation in different species and provided new mechanistic insights into the AKR2 interactome. The combined scientific and artistic perspectives resulted in a multidisciplinary approach advancing our knowledge on AKR/SUB interactome and provided new insights into the function of AKR2-protein interactions in the regulation of the NF-kB pathway. Additionally, herein we proposed an algorithm for quantum vaccinomics by focusing on the model proteins AKR/SUB. This study was recently accepted for publication in Vaccines (Artigas-Jerónimo, S., Pastor Comín, J.J., Villar, M., Contreras, M., Alberdi, P., León Viera, I., Soto, L., Cordero, R., Valdés, J.J., Cabezas-Cruz, A., Estrada-Peña, A., de la Fuente, J. 2020. A novel combined scientific and artistic approach for advanced characterization of interactomes: the Akiri/Subolesinn model. Vaccines 8, 77)
1. Introduction
Biology and art have collaborated before for communication in areas such as human physiology and evolution [1][2][3][4], even proposing that art may have biological basis[5]. As scientists, art reminds us of the power of curiosity, which frequently gets lost during research, and ask questions that are relevant for the investigation, thus supporting that science benefits when artists get involved in research[6][7]. Art visual and musical representations translate into complex or unrecognized concepts and provide a way to better understand and approach scientific challenges[8][10]. Art also contributes to highlighting social concerns addressed by scientists[4]. In this way, art and science converge in the purpose of moving society forward and both benefit from the learning process[9][10][11].
In this study, we provided a perspective of the combined scientific and artistic approach to the interactome using as a model the regulatory protein Akirin, from the Japanese “akiraka ni suru” meaning “making things clear” (AKR; also known as Subolesin (SUB) in ticks). To address this question, herein we proposed a novel methodology to approach challenges in molecular biology using as a model the characterization of the AKR2 interactome and its functional role in the regulation of the NF-kB signaling pathway in human placenta cells. The results of this approach advanced our knowledge in this area by showing that AKR/SUB protein dimerization/multimerization and the physical interactions with different proteins simultaneously are involved in the regulation of various biological processes defined by cell type-specific AKR2-protein interactions and the role of these interactions in the positive and negative regulation of the NF-kB pathway. Additionally, the characterization of the AKR2 interactome may have implications in quantum vaccinomics as a new approach for the development of vaccines for the control of vector infestations and infection/transmission of vector-borne pathogens.
Our combined scientific and artistic perspectives provided a multidisciplinary complementary approach to advance the knowledge of the AKR/SUB model regulatory factor and supported the collaboration between science and art in research (Table 1).
Table 1. Highlights of the collaboration between artists and scientists in this study.
|
Artist contribution |
Scientist perspective |
|
Methodological approach |
|
|
Three visual artists and a musician were invited to read a simplified version of our recent review on AKR/SUB functional evolution [12] and were challenged with the conserved function of these proteins in the regulation of different biological processes throughout the metazoan. |
In response to this challenge, the artists contributed the pieces, musical scores and interpretations shown in the paper to provide their view on this matter. Artist’s contributions served to inspire scientists to discuss and find new perspectives on unexplored characteristics of these proteins with putative functional implications. |
|
Piece El Beso (The Kiss) by Israel León Viera (mixed media on canvas, 2018, 150 x 133 cm; dedicated to his daughter Sofía León Jorge; courtesy KGJ Collection, Spain)
|
|
|
The artist represents the origins of life, with multiple geometric images that interact to illustrate in different species the conserved function of these proteins in biological processes represented by the sea, paper boat, Picasso’s dove, and a fetus growing in mother’s womb while opening the eyes to the world. |
From scientist’s perspective a new and unexplored facet of possible functional relevance of AKR dimerization was proposed in this piece. |
|
Piece “La Danza Molecular” (Molecular Dance) by Leandro Soto (mixed media on paper, 2018, 51 x 71 cm; courtesy KGJ Collection, Spain) |
|
|
The artist describes the constant movement of these proteins that translate into the visual rhythm of repetitive interconnected interactions of forms and colors. |
From scientist’s perspective, the possibility that AKR physically interacts with different proteins simultaneously to regulate various biological processes defined by cell-specific AKR-protein interactions was proposed in this piece. |
|
Piece “Nothing wants to say something” by Raúl Cordero (acrylic, polyester, oil and metallic pigments on canvas, 210 x 190 cm; courtesy of the artist)
|
|
|
The artist proposed that what you are looking for is also looking for you, highlighting that nothing wants to tell you something and it is only a matter of finding how to perceive the message. |
From scientist’s perspective this message challenges the view that AKR-protein interactions occur randomly and suggested that these interactions are functionally relevant in the regulation of different biological processes as occurs with other regulatory factors. |
|
Musical scores |
|
|
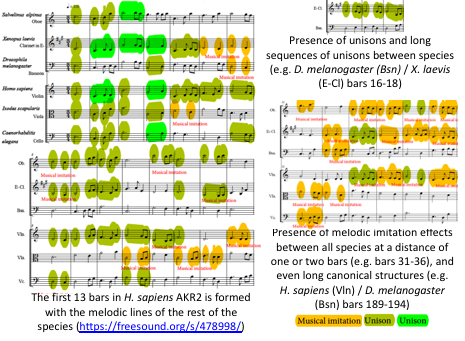
A revised algorithm using musical ensembles was applied to translate into music the AKR and interacting protein sequences. Audio files were uploaded and can be found at https://freesound.org/people/josedelafuente/sounds/478998 to 479009 |
The results supported that AKR in different species are evolutionarily related and structurally conserved and confirmed AKR2-protein interactions identified in this study. Therefore, this tool may be used for evolutionary studies and the prediction of protein-protein interactions. |
Phylogenetic or evolutionary trees are the most accepted approach to illustrate the inferred evolutionary relationships between different species based on the similarities and differences in their physical or genetic characteristics. Furthermore, selective evolutionary pressures have driven a non-simple random-based organization of the genome of surviving organisms. Several methods have been applied to the formulation of DNA language and encoded information, showing that there is at least some degree of organization in the nucleotide sequences found in many organisms[8][13][14][15]. Our results advanced a step forward the use of the DNA language approach by characterizing not only evolutionary relationships but also functional protein interactions (Table 1).
The results of this study provided a novel combined scientific and artistic multidisciplinary approach to address challenging questions in molecular biology. The collaboration between scientists and artists provided two main methodological outcomes: (a) the suggestion by visual artists of the scientific characterization of previously unexplored properties of AKR/SUB and (b) the application of an algorithm using musical ensembles based on AKR/SUB and interacting protein sequences as a new method to predict protein-protein interactions (Table 2). The recent review of AKR/SUB functional evolution[12] suggested that these proteins evolved with conserved sequence, structure and function, and the need to further characterize its function in different conserved biological pathways such as NF-kB. The need to better understand AKR/SUB function was addressed in this study and although further experiments are required to fully characterized the role of the AKR/SUB interactome in the regulation of the NF-kBpathway, the results in human placenta cells served as a methodological proof-of-concept to advance this research area. The results showed that AKR-AKR interactions result in protein dimerization/multimerization with possible functional implications that require attention. AKR/SUB physically interacts with different proteins simultaneously to regulate various biological processes with cell type differences in AKR2 interactome that are likely associated with the specific biological processes regulated by this protein in each cell type. In this way, multiple interactions between AKR2 and interacting proteins differentially regulate the NF-kB pathway by affecting gene expression controlled by NF-kB in a biological context-dependent manner. In addition to cell type-specific differences in AKR/SUB-protein interactions, species-specific differences have been also described and require further attention[12]. For example, it has been shown in tick cells that SUB regulome affects multiple biological processes that vary in response to infection with the tick-borne pathogen, Anaplasma phagocytophilum[12]. Therefore, studies in other cells such as lymphocytes or monocytes and promyelocytic leukemia HL-60 model cells are required to evaluate cell type-specific differences in the function of AKR2-protein interactions in NF-kB signaling. Functionally, AKR2-protein interactions in human placenta cells differentially regulate the NF-kB signaling pathway, suggesting that AKR2 interacting proteins might constitute suitable secondary transcription factors for cell and stimulus-specific regulation of NF-kB. As shown previously by genetic approaches, musical ensembles supported that AKR/SUB in different species are evolutionarily related and structurally conserved and predicted AKR2-protein interactions coinciding with those found by genetic approaches such as the Y2H screening used here. The methodological approach proposed here for quantum vaccinomics further advances the potential of the vaccinomics pipeline used before for the identification of candidate protective antigens[16][17][18]. Furthermore, if combined with network analysis for the integration of interactomics and regulomics datasets[19] and Big Data machine learning algorithms to identify candidate protective antigens and epitopes[20], quantum vaccinomics would result in designing chimeric antigens based on protective epitopes in SID from vector and pathogen derived regulatory proteins. Vaccines with these chimeric protective antigens would address the possibility of effective and sustainable control of vector-borne diseases by targeting multiple ectoparasite and pathogen species in multiple hosts. The collaboration between scientists and artists from multiple disciplines has a positive impact on advancing research to address scientific challenges.
References
- Jeffries, S. When two tribes meet: collaborations between artists and scientists. The Guardian 2011, August 21.
- Available online: https://www.theguardian.com/artanddesign/2011/aug/21/collaborations-between-artists-and-scientists
- Tayag, Y.; Wells, B. Art and evolution: a work in progress. Scientific American 2014, May 6. Available online: https://blogs.scientificamerican.com/guest-blog/art-and-evolution-a-work-in-progress/
- Schulkin, J.; Raglan, G.B. The evolution of music and human social capability. Front. Neurosci. 2014, 8, 292.
- de la Fuente, J.; Estrada-Peña, A.; Cabezas-Cruz, A.; Brey, R. Flying ticks: anciently evolved associations that constitute a risk of infectious disease spread. Parasit. Vectors 2015, 8, 538.
- Veis, N. Four takes in the evolution of art. Nature 2017, 543, 490.
- Stevens, C.; O´Connor, G. When artists get involved in research, science benefits. The Conversation 2017, August 16. Available online: http://theconversation.com/when-artists-get-involved-in-research-science-benefits-82147
- de la Fuente, J. Anaplasmosis: What we can learn from Lam´s surrealistic animalarium. Hektoen International 2018, Hektorama-Infectious Diseases-Summer 2018. Available online: http://hekint.org/2018/08/23/anasplasmosis-what-we-can-learn-from-lams-surrealistic-animalarium/. See also other publications in Hektoen International (http://hekint.org).
- Riego, E.; Silva, A.; de la Fuente, J. The sound of the DNA language. Biol. Res. 1995, 28, 197-204.
- Eldred, S.M. Art–science collaborations: Change of perspective. Nature 2016, 537, 125-126.
- Shelton, J. A visual artist compares the way scientists and artists see a world of discovery. YaleNews 2017, April 3. Available online: https://news.yale.edu/2017/04/03/visual-artist-compares-way-scientists-and-artists-see-world-discovery
- Maeda, J. Artists and scientists: More alike than different. Scientific American 2013, July 11. Available online: https://blogs.scientificamerican.com/guest-blog/artists-and-scientists-more-alike-than-different/
- Artigas-Jerónimo, S.; Villar, M.; Cabezas-Cruz, A.; Valdés, J.J.; Estrada-Peña, A.; Alberdi, P.; de la Fuente, J. Functional evolution of Subolesin/Akirin. Front. Physiol. 2018, 9, 1612.
- Ailenberg, M.; Rotstein, O. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques 2009, 47, 747-754.
- Temple, M.D. An auditory display tool for DNA sequence analysis. BMC Bioinformatics 2017, 18, 221.
- Mannone, M. Knots, music and DNA. J. Creat. Music Syst. 2018, 2, doi:https://doi.org/10.5920/jcms.2018.02
- de la Fuente, J.; Kopáček, P.; Lew-Tabor, A.; Maritz-Olivier, C. Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol. 2016, 38, 754-769.
- de la Fuente, J.; Merino, O. Vaccinomics, the new road to tick vaccines. Vaccine 2013, 31, 5923–5929.
- Contreras, M.; Villar, M.; de la Fuente, J. A vaccinomics approach to the identification of tick protective antigens for the control of Ixodes ricinus and Dermacentor reticulatus infestations in companion animals. Front. Physiol. 2019, 10, 977.
- Artigas-Jerónimo, S.; Estrada-Peña, A.; Cabezas-Cruz, A.; Alberdi, P.; Villar, M.; de la Fuente, J. Modeling modulation of the tick regulome in response to Anaplasma phagocytophilum for the identification of new control targets. Front. Physiol. 2019, 10, 462.
- de la Fuente, J.; Villar, M.; Estrada-Peña, A.; Olivas, J.A. High throughput discovery and characterization of tick and pathogen vaccine protective antigens using vaccinomics with intelligent Big Data analytic techniques. Expert Rev. Vaccines 2018, 17, 569-576.