Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sonali Bhakta | -- | 2271 | 2022-09-30 02:59:22 | | | |
| 2 | Rita Xu | -14 word(s) | 2257 | 2022-09-30 03:09:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Bhakta, S.; Tsukahara, T. C-to-U RNA Editing. Encyclopedia. Available online: https://encyclopedia.pub/entry/28076 (accessed on 08 February 2026).
Bhakta S, Tsukahara T. C-to-U RNA Editing. Encyclopedia. Available at: https://encyclopedia.pub/entry/28076. Accessed February 08, 2026.
Bhakta, Sonali, Toshifumi Tsukahara. "C-to-U RNA Editing" Encyclopedia, https://encyclopedia.pub/entry/28076 (accessed February 08, 2026).
Bhakta, S., & Tsukahara, T. (2022, September 30). C-to-U RNA Editing. In Encyclopedia. https://encyclopedia.pub/entry/28076
Bhakta, Sonali and Toshifumi Tsukahara. "C-to-U RNA Editing." Encyclopedia. Web. 30 September, 2022.
Copy Citation
The restoration of genetic code by editing mutated genes is a potential method for the treatment of genetic diseases/disorders. Genetic disorders are caused by the point mutations of thymine (T) to cytidine (C) or guanosine (G) to adenine (A), for which gene editing (editing of mutated genes) is a promising therapeutic technique. In C-to-Uridine (U) RNA editing, it converts the base C-to-U in RNA molecules and leads to nonsynonymous changes when occurring in coding regions; for G-to-A mutations, A-to-I editing occurs. Editing of C-to-U is not as physiologically common as that of A-to-I editing.
RNA editing 1
cytidine 2
thymine 3
uridine 4
mutation 5
1. Introduction
RNA editing is a biological process or tool for repairing or altering RNA in a mitochondrion-encoded mRNA of a kinetoplastid trypanosome. RNA editing was first introduced to describe a process that occurs in trypanosomes and involves the insertion and deletion of uridine monophosphate (UMP) inside nascent transcripts after transcription [1]. Since the discovery of the post-transcriptional sequence, the number of techniques associated with the term RNA editing has grown. The insertion and deletion of nucleotides other than UMP, base deamination, and the co-transcriptional insertion of non-template nucleotides are now referred to as RNA editing. RNA editing has been observed in mRNAs, tRNAs, and rRNAs, in mitochondrial and chloroplast encoded RNAs, as well as in nuclear encoded RNAs [1]. Examples of RNA editing have been found in many Metazoa, unicellular eukaryotes, such as trypanosomes, and plants. RNA editing has been observed in prokaryotes on a small scale although some researchers have made detailed study on the tRNA editing in E. coli [2].
RNA editing tools are basically categorized into two types depending on their response mechanisms. For example, insertion/deletion RNA editing, involves the insertion or deletion of targeted nucleotides with the aim of changing the codon sequence of the targeted mRNA [1]. However, this RNA editing can be done in another way as well, where it turns/alters one encoded nucleotide into a new nucleotide via base alteration or modification without modifying the overall length of the RNA. As a result, the codon sequence is ultimately changed; this is particularly used for the treatment of single nucleotide mutations.
2. RNA Editing
RNA editing as a therapeutic approach was first conceptualized and utilized as a therapy in 1995. The main purpose of this method is to restore RNA sequences in order to treat genetic diseases caused by point mutations. Advanced research has enhanced and established this technology, which is now known as artificial site-directed RNA editing for restoring RNA. This unique therapeutic approach has the potential to be utilized to cure diseases, such as numerous neurological maladies in humans, by restoring the mutated A or C in mRNA without changing or affecting the genome sequence of the mRNA target [3]. A-to-I and C-to-U editing are two types of substitutional RNA editing in mammals. Due to the higher potential of recoding point mutations, many studies have focused on changing as well as imitating RNA editing. RNA editing of C-to-U is commonly found among flowering plants and mainly occurs within mitochondrial protein regions with highly conserved amino acid sequences [4].
3. C-to-U RNA Editing
RNA editing in C-to-U is a process or therapeutic approach that converts a single or multiple C-to-U nucleotides in transcript sequences. C-to-U RNA editing can generate start or stop codons that can change the encoded amino acids depending on preferences towards the splice site [5]. The C-to-U type of RNA editing was originally illustrated in vertebrates for apolipoprotein B (apoB) encoding mRNA. Deamination through hydrolysis at the C4 site of cytidine (C) was later found to be involved in apoB editing [6][7]. The presence of both cis-acting elements (tripartite regulatory sequences) and trans-acting elements around the altered cytidine is required for this conversion or editing (the editosome is a multiprotein complex that contains a catalytic cytidine deaminase and many auxiliary proteins) [3]. Editing of C-to-U at the RNA level has been found in higher family plants, particularly in the mitochondria and chloroplasts [8].
4. Artificial C-to-U RNA Editing
Both C-to-U and A-to-I conversions are included in enzymatic site-directed RNA editing. Recently, artificial site-directed RNA editing of A-to-I has been successfully carried out in vitro and in cells as well as in vivo [8][9]. However, few reports of artificial site-directed C-to-U RNA editing have been published recently. The RNA editing machinery relies on two critical components: complementary RNA sequences that can precisely bind to specified sequences (guide RNA) and deamination-editing enzyme/editors. Furthermore, non-enzymatic site-directed C-to-U editing, which does not have the same constraints as site-directed enzymatic RNA editing, was recently identified and has generated a lot of interest in this field of research. Researchers have focused on C-to-U RNA editing with special emphasis on the enzymatic approach [8].
5. Enzymes (Editors) for Artificial C-to-U RNA Editing
The artificial or enzymatic approach of deamination from C-to-U is mainly dependent on the enzymes from the apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) family proteins. Eleven genes code for members of the APOBEC family that have been discovered to date (APOBEC1, APOBEC2, APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, APOBEC3H, APOBEC4, and AICDA/AID). They all have a zinc-dependent deaminase domain (ZDD) [10][11]. Among all of these APOBEC subfamily proteins only APOBEC-1, 3A, 3 B, and 3G (Figure 1) have been proven to mediate the C-to-U RNA editing [12][13][14][15]. APOBEC-1 was the first member of the APOBEC family to be discovered and researched, and its significance in apolipoprotein B (ApoB) mRNA editing has been well documented.
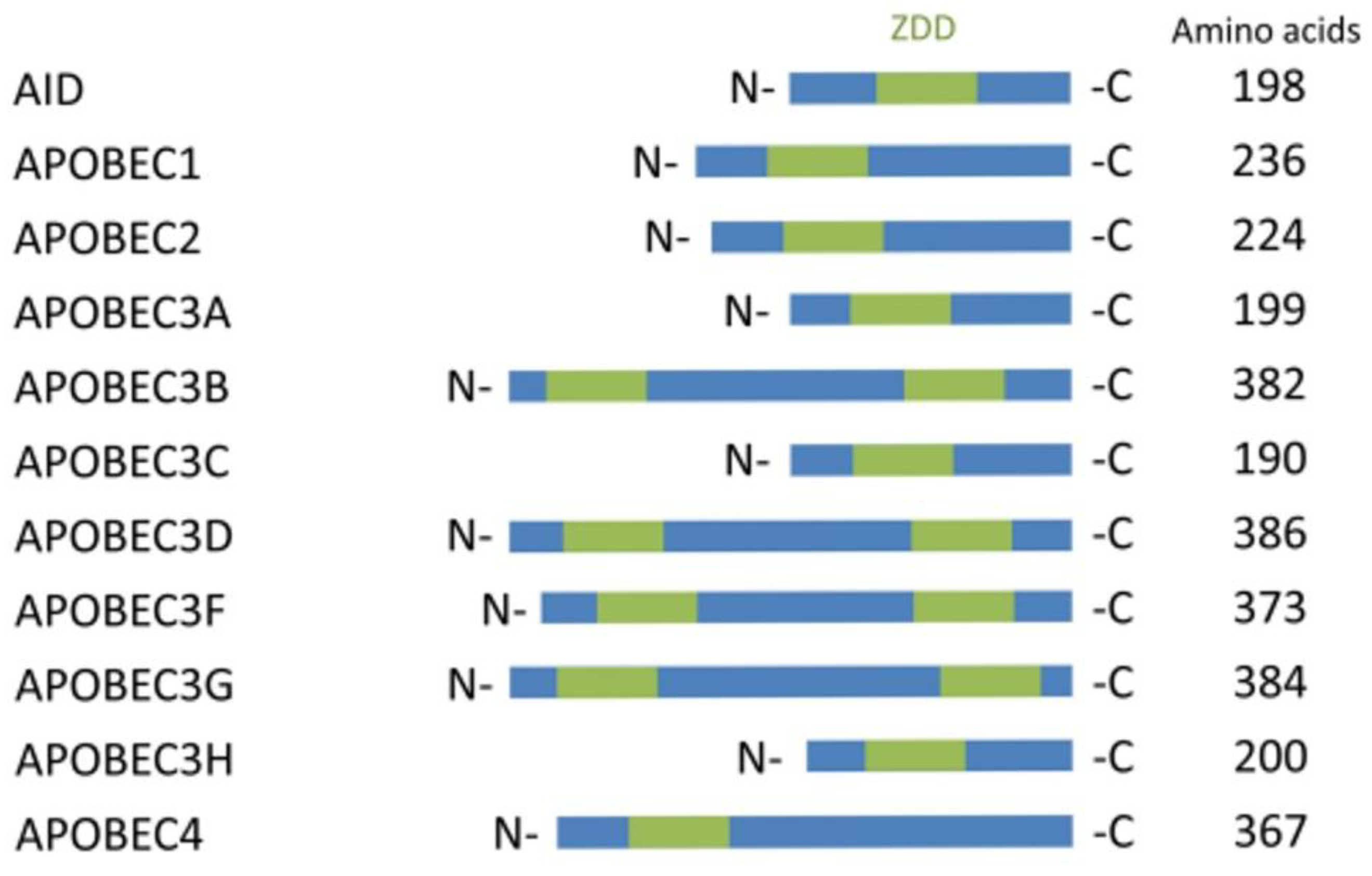
Figure 1. APOBEC family proteins for C to U editing.
For the enzymatic approach, the MS2 system along with the APOBEC family protein enzyme has been a very promising technique for the therapeutic RNA editing technique. C-to-U editing (Figure 2) using the MS2 system (MS2 stem loop along with MS2 coat protein) and APOBEC1 has been previously performed by Bhakta et al. [16] by converting BFP (Blue Fluorescence Protein having a single mutated T-to-C) to GFP (Green Fluorescence Protein- which is restored after the editing from C-to-U) (Figure 3).
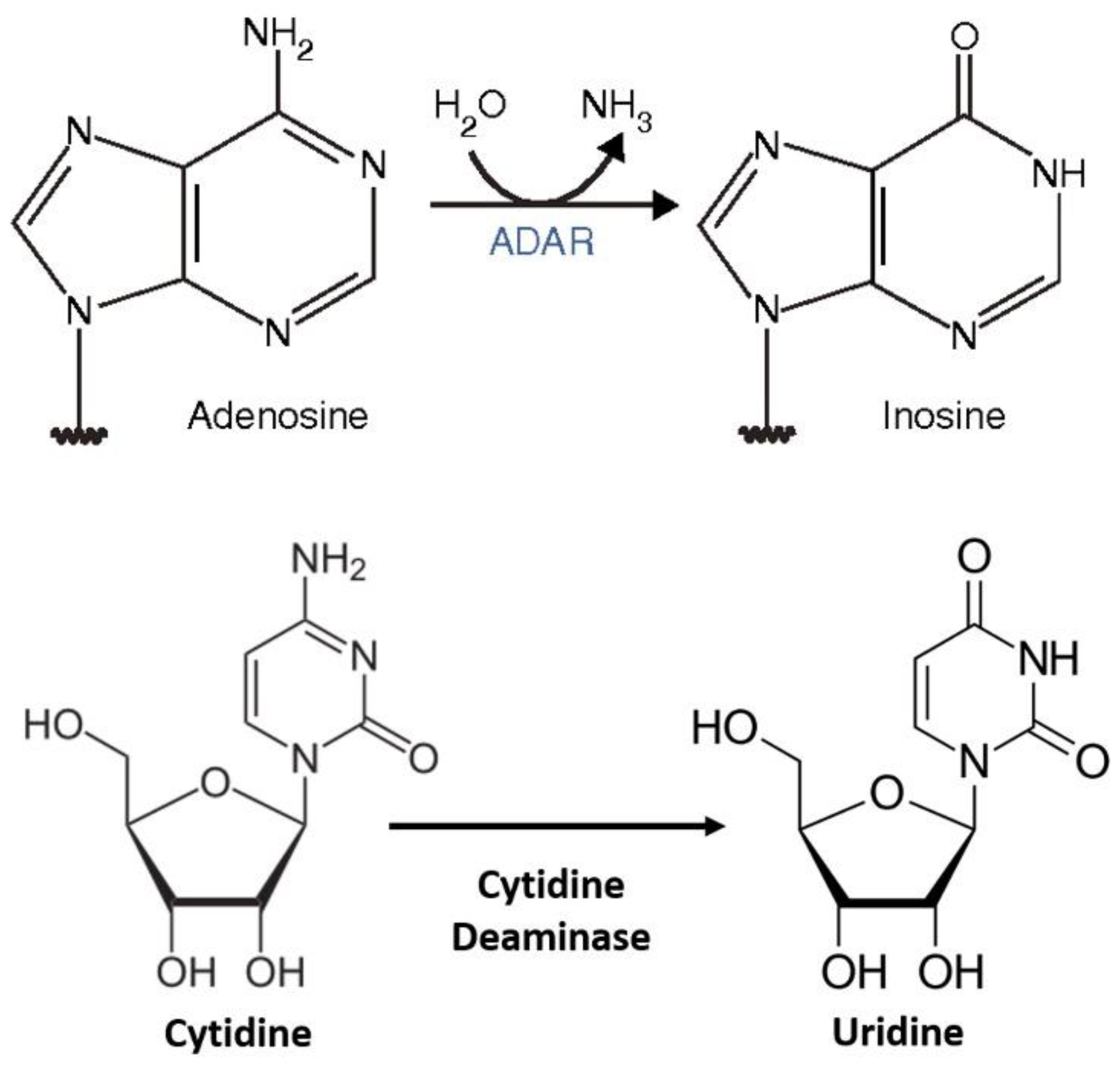
Figure 2. RNA editing (A to I and C to U) [17].
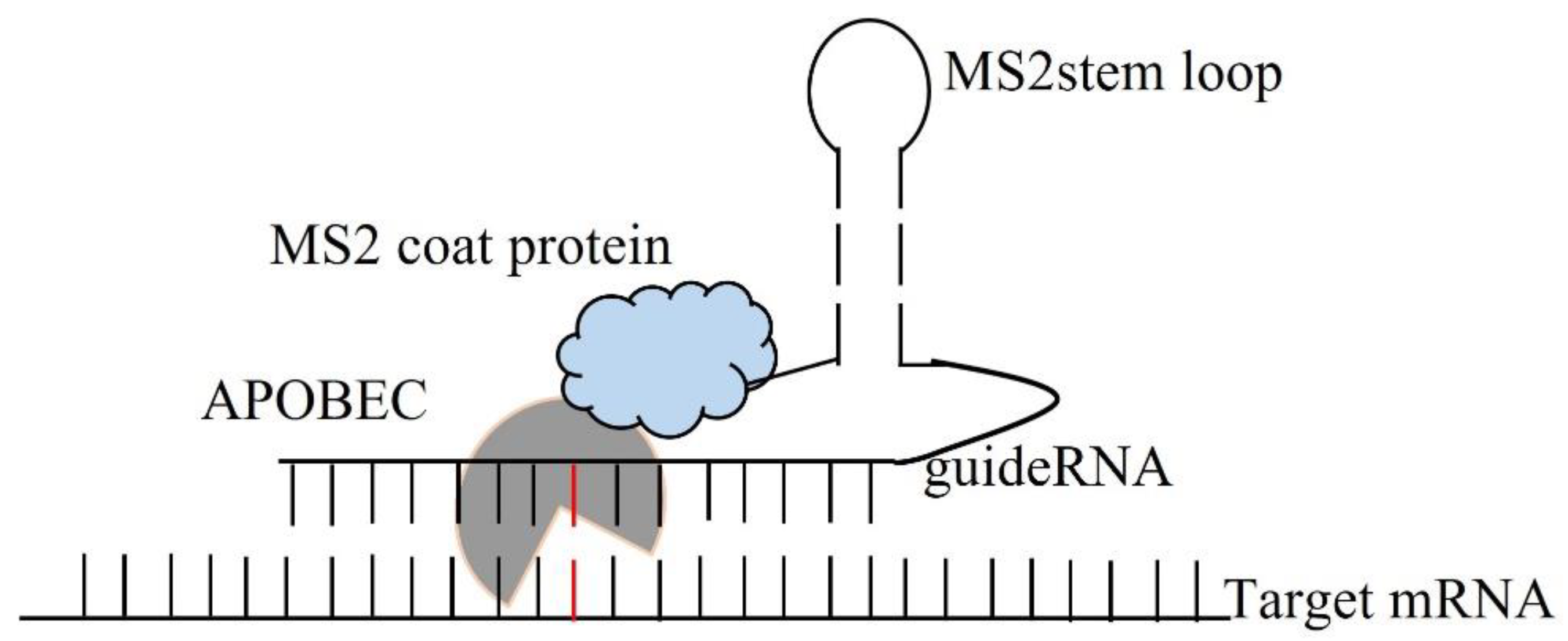
Figure 3. Enzymatic C to U RNA editing by APOBEC 1 deaminase.
Only vertebrates have APOBECs, and this type of RNA editing is the second most common after ADAR (Adenosine Deaminase acting on RNA) editing. Unlike ADARs, the APOBEC family of proteins, including the Alu (Arthrobacter luteus) sequence, primarily affects non-coding and intronic sequences [18][19]. Surprisingly, the APOBEC family of proteins are not just for RNA editing. They were first introduced as tools for the editing of single-stranded DNA (ssDNA) and genomic DNA (gDNA), respectively. As a result, APOBEC-mediated DNA editing has received more attention and is better understood than APOBEC-mediated RNA editing. The efficiency of genome/DNA editing cannot be compared to RNA editing, however, because the deamination of C in DNA results in U; APOBEC-mediated DNA editing can result in the degradation of viral DNA, resulting in a reduction of virus replication or multiplication. Moreover, uracil-rich viral DNA can trigger DNA damage and stress–response pathways, causing natural killer (NK) cells to up-regulate activating ligands (NKG2D ligands) and destroy infected cells [15].
The APOBEC family of proteins plays a vital role in the introduction of mutations in cancerous tissues [20][21][22][23][24]. These mutations are primarily caused by genome/DNA editing or abnormal APOBEC enzyme production. DNA editing mediated by APOBECs for C-to-U editing has been thoroughly characterized by Knisbacher et al. [23]. DNA editing aids in the natural mechanisms of the body. However, DNA or genome editing is essential for a good and efficient adaptive immune response. Somatic hypermutation is the most commonly known example, occurring in sequences encoding hypervariable portions of immunoglobulins, which result in the formation of high-affinity antibodies [19].
ApoB-100, the full-length form of ApoB protein, is expressed in hepatic cells (hepatocytes) in the liver. APOBEC-1 RNA editing, on the other hand, causes an early stop codon in ApoB mRNA in the small intestine, resulting in the premature termination of translation. Consequently, another isoform of ApoB-48 was created. The full-length form (ApoB-100) carries cholesterol in the bloodstream, whereas the shortened form (ApoB-48) transports triglycerides [25].
There are seven APOBEC-3 paralogs in the human genome (APOBEC-3A, 3 B, 3C, 3D, 3F, 3G, and 3H). Although all of these paralogs bind to RNA [26], only three of them have shown RNA editing activity (APOBEC-3A, APOBEC-3B, and APOBEC-3G). These roles have an impact on the immune system. Under hypoxic conditions and IFN activation, they were found to be expressed in macrophages, monocytes, and NK cells [27][28][29][30]. Furthermore, they are expressed in human natural Tregs in response to CD3/CD28 stimulation, especially APOBEC-3G, 3D, and 3H [30]. Single gene encoded APOBEC-3 was identified for the first time as the Friend leukemia virus resistance (Fvr) gene [31] in mice. Mouse APOBEC3 is most similar to human APOBEC3G and contributes to viral resistance by mutating viral DNA. In recent times the CRISPR-Cas9-APOBEC editing system has had a significant impact on DNA editing, but off-target effects are a major concern in this case [31].
6. C-to-U RNA Editing in Mammals
The restoration of genetic code from A-to-I and C-to-U are the two most common RNA editing processes in mammalian cells [32][33]. Deamination has been reported to occur from A-to-I in hundreds of thousands of places, with the most common occurring in intronic and non-coding regions, notably with Alu sequence repeated targets [34][35]. A-to-I RNA editing in coding domains is frequently incorporated with brain proteins that can recode [30]. However, C-to-U editing is less commonly found in humans than is A-to-I [36]. The pre-mRNA of apolipoprotein B (apoB) is predominantly located in intestinal cells, and currently approved targets in mRNA are among the physiologically minimum recognized C-to-U RNA editing targets. In an earlier investigation, previously unknown 32 APOBEC1 (apoB editing catalytic subunit 1) editing sites were identified in the 3′-untranslated regions (3′-UTRs) of diverse mRNA transcripts [37]. Furthermore, in the AU-rich parts of the 3′-UTRs, 56 novel modifying sites were identified, among which 54 were intestinal mRNAs, within which 22 unique editing points were discovered in mRNAs of the liver [38]. In macrophages derived from bone marrow, 410 C-to-U RNA editing events were found, among which 97% of C-to-U events were found to occur in 3′-UTRs [39]. Moreover, C-to-U RNA editing events of apoB pre-mRNA occur in the nucleus [40][41]. At the C6666U editing site, the conversion from glutamine (CAA) to a stop codon (UAA) occurs at the in-frame translational site. The ApoB48 protein is produced by C6666U-edited apoB RNA, whereas ApoB100 protein is produced by C6666-unedited apoB RNA [40].
In the C6802U editing site, the threonine codon (ACA) is changed to an isoleucine (AUA). Because the C6802U editing event occurs concurrently with the C6666U editing event, C6802U is not expressed in the truncated ApoB48 protein but in the mRNA [38]. The RNA editing of C-to-U is necessary for the stoichiometric modulation of trans-acting components within the macromolecular enzyme complex (editosome), which is responsible for targeted deamination. These cis-acting elements, in combination with trans-acting factors, are required for C-to-U RNA editing in vitro. They are made up of 50 nucleotides modifying the edited cytidine that contains a regulatory tripartite motif, which contains an 11-nt motif (UGAUCAGUAUA) located in a sequence (the mooring sequence) downstream of the edited base [42][43][44][45][46]. To generate a stable secondary structure that increases specificity, the 3′ mooring sequence is combined with a 5′ efficiency sequence [44][45].
The editosome of C-to-U editing consists of a minimum of three protein components: ApoB1 and two essential cofactors, ApoB1-complementary factor (ACF) and RNA binding motif 47 (RBM47) [46][47]. The cytidine deaminases (RNA-specific) APOBEC family includes APOBEC1. Like the cytidine deaminase family-derived members, APOBEC1 possesses a zinc-dependent deaminase domain that is essential for the deamination of C [48][49]. In the catalytic domain of APOBEC1, specific amino acids are bound to AU-rich areas in apoB pre-mRNA, producing homodimers. In vitro, this interaction is inadequate for mRNA association, which needs to be used as a cofactor of ACF, a potential RNA-binding protein (RBP). In vitro experiments revealed that this cofactor has a high affinity for the mooring sequence and forms a minimal editosome with APOBEC1 [50]. Elav/HelN1/HuR is a protein that consists of an RNA-recognition motif (RRM) of single-stranded RNA, repeated several times. The N- and C-terminal areas bordering many RRMs are required for the interaction of ACF with the APOBEC1 enzyme [51].
While ACF knockout animals may die during early pregnancy, ACF+/mutant mice show a higher editing efficiency, contradicting the idea that cofactors are essential for editosome editability in vitro. Despite the abundance of scientific evidence for cofactors and C-to-U APOBEC-derived deaminase editing in vitro, there is no strong proof that cofactors are essential for C to-U RNA editing in vivo. As a result, the function of cofactors (ACF) in vivo remains unclear [50]. C-to-U RNA editing in vivo requires an extra cofactor, which has recently been discovered as RBM47 [50]. In the holoenzyme of the editosome, RBM47 interacts with APOBEC1 and ACF, and works with APOBEC1 to edit transcripts of ApoB. However, the consequences of the ACF-RBM47 interaction in vivo are not yet fully known. In vitro, RBM47 can also play the role of ACF cofactors in the RNA editing of the C-to-U enzyme complex. The C-to-U RNA editing of apoB and four other C-to-U RNA editing targets (Sult1d1—sulfotransfer RBM47) is an editosome component that is essential for C-to-U RNA editing [52]. A novel enzyme for C-to-U RNA editing has been identified as APOBEC3A (A3A) (Figure 4), DYW, a structurally related member of the cytidine deaminase family, which is expressed mostly in myeloid cells such as macrophages and monocytes [53][54].
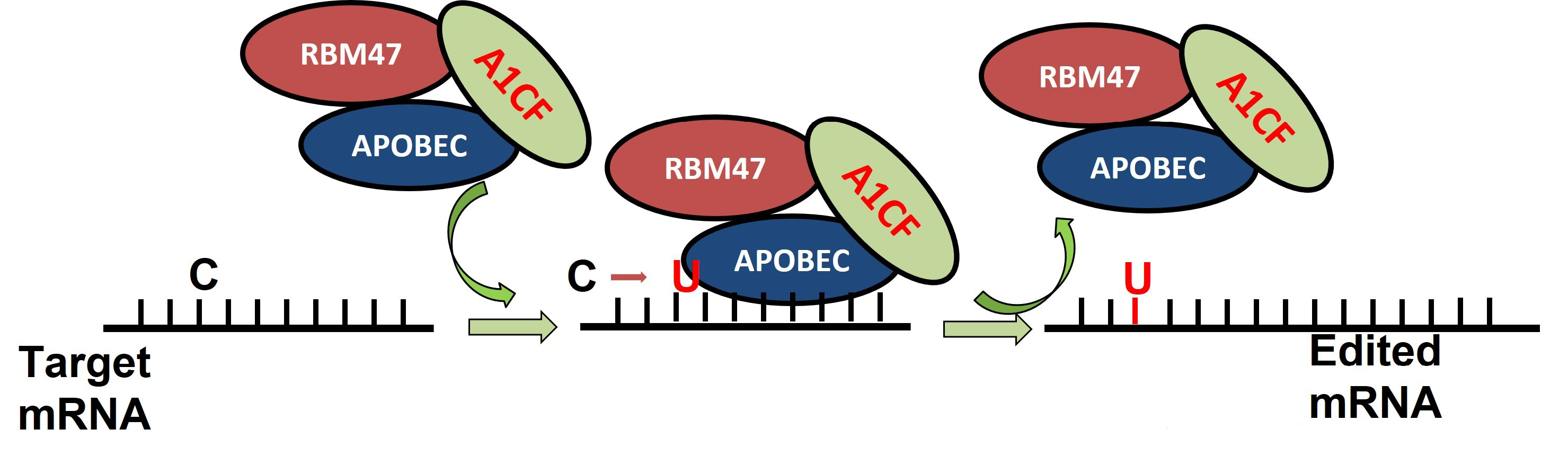
Figure 4. RNA-binding protein RBM47 is required for normal Cytidine to Uridine RNA editing in mammals and is sufficient for the C to U RNA editinf activity of APOBEC demainase domain [51].
References
- Wolf, J.; Gerber, A.P.; Keller, W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002, 21, 3841–3851.
- Brennicke, A.; Marchfelder, A.; Binder, S. RNA editing. FEMS Microbiol. Rev. 1999, 23, 297–316.
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349.
- Zanlungo, S.; Béqu, D.; Quiñones, V.; Araya, A.; Jordana, X. RNA editing of a pocytochrome b (cob) transcripts in mitochondria from two genera of plants. Curr. Genet. 1993, 24, 344–348.
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531.
- Chen, S.H.; Habib, G.; Yang, C.Y.; Gu, Z.W.; Lee, B.R.; Weng, S.A.; Silberman, S.R.; Cai, S.J.; Deslypere, J.P.; Rosseneu, M.; et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific inframe stop codon. Science 1987, 238, 363–366.
- Powell, L.M.; Wallis, S.C.; Pease, R.J.; Edwards, Y.H.; Knott, T.J.; Scott, J. A novel form of tissue-specific RNA processing produces a polipoprotein-B48 in intestine. Cell 1987, 50, 831–840.
- Freyer, R.; Kiefer-Meyer, M.C.; Kössel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA 1997, 94, 6285–6290.
- Bahn, J.H.; Lee, J.H.; Li, G.; Greer, C.; Peng, G.; Xiao, X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012, 22, 142–150.
- Navaratnam, N.; Bhattacharya, S.; Fujino, T.; Patel, D.; Jarmuz, A.L.; Scott, J. Evolutionary origins of apoB mRNA editing: Catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 1995, 81, 187–195.
- Blanc, V.; Davidson, N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 594–602.
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881.
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.F. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016, 6, 39100.
- Patnaik, S.K.; Kannisto, E. APOBEC3B is a new RNA editing enzyme. In Proceedings of the RNA 2016, Annual Meeting of RNA Society, Kyoto, Japan, 28 June–2 July 2016.
- Moris, A.; Murray, S.; Cardinaud, S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014, 5, 534.
- Bhakta, S.; Sakari, M.; Tsukahara, T. RNA editing of BFP, a point mutant of GFP, using artificial APOBEC1 deaminase to restore the genetic code. Sci. Rep. 2020, 10, 17304.
- Gerber, A.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384.
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976.
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC Enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015, 5, 704–712.
- Kung, C.; Maggi, L.B., Jr.; Weber, J.D. The Role of RNA Editing in Cancer Development and Metabolic Disorders. Front. Endocrinol. 2018, 9, 762.
- Chan, K.; Roberts, S.A.; Klimczak, L.J.; Sterling, J.F.; Saini, N.; Malc, E.P.; Kim, J.; Kwiatkowski, D.J.; Fargo, D.C.; Mieczkowski, P.A.; et al. An APOBEC3A hyper-mutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015, 47, 1067–1072.
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370.
- Knisbacher, B.A.; Gerber, D.; Levanon, E.Y. DNA Editing by APOBECs: A genomic preserver and transformer. Trends Genet. 2016, 32, 16–28.
- Davidson, N.O.; Shelness, G.S. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu. Rev. Nutr. 2000, 20, 169–193.
- Prohaska, K.M.; Bennett, R.P.; Salter, J.D.; Smith, H.C. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip. Rev. RNA 2014, 5, 493–508.
- Sharma, S.; Patnaik, S.K.; Kemer, Z.; Baysal, B.E. Transient overexpression of exogenous APOBEC3A causes C to U RNA editing of thousands of genes. RNA Biol. 2017, 14, 603–610.
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Cortes Gomez, E.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019, 20, 37.
- Marek-Trzonkowska, N.; Piekarska, K.; Filipowicz, N.; Piotrowski, A.; Gucwa, M.; Vogt, K.; Sawitzki, B.; Siebert, J.; Trzonkowski, P. Mild hypothermia provides Treg stability. Sci. Rep. 2017, 7, 11915.
- Takeda, E.; Tsuji-Kawahara, S.; Sakamoto, M.; Langlois, M.A.; Neuberger, M.S.; Rada, C.; Miyazawa, M. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J. Virol. 2008, 82, 10998–11008.
- Lai, F.; Drakas, R.; Nishikura, K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 1995, 270, 17098–17105.
- McGrath, E.; Shin, H.; Zhang, L.; Phue, J.N.; Wu, W.W.; Shen, R.F.; Jang, Y.Y.; Revollo, J.; Ye, Z. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat. Commun. 2019, 10, 5353.
- Smith, H.C.; Bennett, R.P.; Kizilyer, A.; McDougall, W.M.; Prohaska, K.M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012, 23, 258–268.
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A to I RNA editing occurs at over a hundred million genomic sites located in a majority of human genes. Genome Res. 2013, 24, 365–376.
- Ramaswami, G.; Lin, W.; Piskol, R.; Tan, M.H.; Davis, C.; Li, J.B. Accurate identification of human alu and non-alu RNA editing sites. Nat. Methods 2012, 9, 579–581.
- Li, J.B.; Church, G.M. Deciphering the functions and regulation of brain-enriched A to I RNA editing. Nat. Neurosci. 2013, 16, 1518–1522.
- Rosenber, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236.
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Hamidane, H.B.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C to U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79.
- Harjanto, D.; Papamarkou, T.; Oates, C.J.; Rayon-Estrada, V.; Papavasiliou, F.N.; Papavasiliou, A. RNA editinggenerates cellular subsets with diverse sequence within populations. Nat. Commun. 2016, 7, 12145.
- Blanc, V.; Davidson, N.O. Mouse and other rodent models of C to U RNA editing. Methods Mol. Biol. 2011, 718, 121–135.
- Navaratnam, N.; Patel, D.; Shah, R.R.; Greeve, J.C.; Powell, L.M.; Knott, T.J.; Scott, J. An additional editing site is present in apolipoprotein B mRNA. Nucleic Acids Res. 1991, 19, 1741–1744.
- Backus, J.W.; Smith, H.C. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992, 20, 6007–6014.
- Backus, J.W.; Smith, H.C. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991, 19, 6781–6786.
- Hersberger, M.; Innerarity, T.L. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J. Biol. Chem. 1998, 273, 9435–9442.
- Nakamuta, M.; Tsai, A.; Chan, L.; Davidson, N.O.; Teng, B.B. Sequence elements required for apolipoprotein B mRNA editing enhancement activity from chicken enterocytes. Biochem. Biophys. Res. Commun. 1999, 254, 744–750.
- Shah, R.R.; Knott, T.J.; Legros, J.E.; Navaratnam, N.; Greeve, J.C.; Scott, J. Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem. 1991, 266, 16301–16304.
- Mehta, A.; Kinter, M.T.; Sherman, N.E.; Driscoll, D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell Biol. 2000, 20, 1846–1854.
- Bhattacharya, S.; Navaratnam, N.; Morrison, J.R.; Scott, J.; Taylor, W.R. Cytosine nucleoside/nucleotide deaminases and apolipoprotein B mRNA editing. Trends Biochem. Sci. 1994, 3, 105–106.
- MacGinnitie, A.J.; Anant, S.; Davidson, N.O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J. Biol. Chem. 1995, 270, 14768–14775.
- Wedekind, J.E.; Dance, G.S.; Sowden, M.P.; Smith, H.C. Messenger RNA editing in mammals: New members of the APOBEC family seeking roles in the family business. Trends Genet. 2003, 19, 207–216.
- Blanc, V.; Henderson, J.O.; Newberry, E.P.; Kennedy, S.; Luo, J.; Davidson, N.O. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol. Cell Biol. 2005, 25, 7260–7269.
- Fossat, N.; Tourle, K.; Radziewic, T.; Barratt, K.; Leibhold, D.; Studdert, J.B.; Power, M.; Jones, V.; Loebel, D.A.F.; Tam, P.P. C to U RNA editing meditated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014, 15, 903–910.
- Liu, Z.; Zhang, J. Human C-to-U Coding RNA Editing Is Largely Nonadaptive. Mol. Biol. Evol. 2018, 35, 963–969.
- Huang, X.; Lv, J.; Li, Y.; Mao, S.; Li, Z.; Jing, Z.; Sun, Y.; Zhang, X.; Shen, S.; Wang, X.; et al. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 2021, 40, e108209.
- Takenaka, M.; Verbitskly, D.; van der Merwe, J.A.; Zehrmann, A.; Brennicke, A. The process of RNA editing in plant mito-chondria. Mitochondrion 2008, 8, 35–46.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Revisions:
2 times
(View History)
Update Date:
30 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




