
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Vasiliki Gkretsi | -- | 2873 | 2022-09-29 08:25:15 | | | |
| 2 | Beatrix Zheng | Meta information modification | 2873 | 2022-09-30 06:20:36 | | | | |
| 3 | Jessie Wu | + 2 word(s) | 2875 | 2022-09-30 07:12:25 | | |
Video Upload Options
LncRNAs are defined as RNAs whose transcript length exceeds 200 nucleotides and is not translated into proteins. They are transcribed by RNA polymerase II and although they can be categorized based on their length, function, location, and mechanism of action, so far there are no standard guidelines for this classification. According to their position in the genome in relation to protein-coding genes, they are categorized as (i) sense, which are transcribed in the same direction as that of the protein-coding gene, (ii) antisense, which are transcribed in a direction opposite to that of the protein-coding gene, (iii) bidirectional, which are transcribed in both directions, (iv) intronic, that are transcribed from introns found within protein-coding genes, (v) intergenic (lincRNAs), that are transcribed between two protein-coding genes, and (vi) enhancer lncRNAs. Small nucleolar RNAs (snoRNAs) are 60–300 nucleotides in length and as the name suggests, are mainly found in the nucleolus where they function as guide RNAs for the post-transcriptional modification of ribosomal RNAs and some spliceosomal RNAs.
1. Long Intergenic Noncoding RNAs (LincRNAs)
2. Small Nucleolar RNAs (snoRNAs)
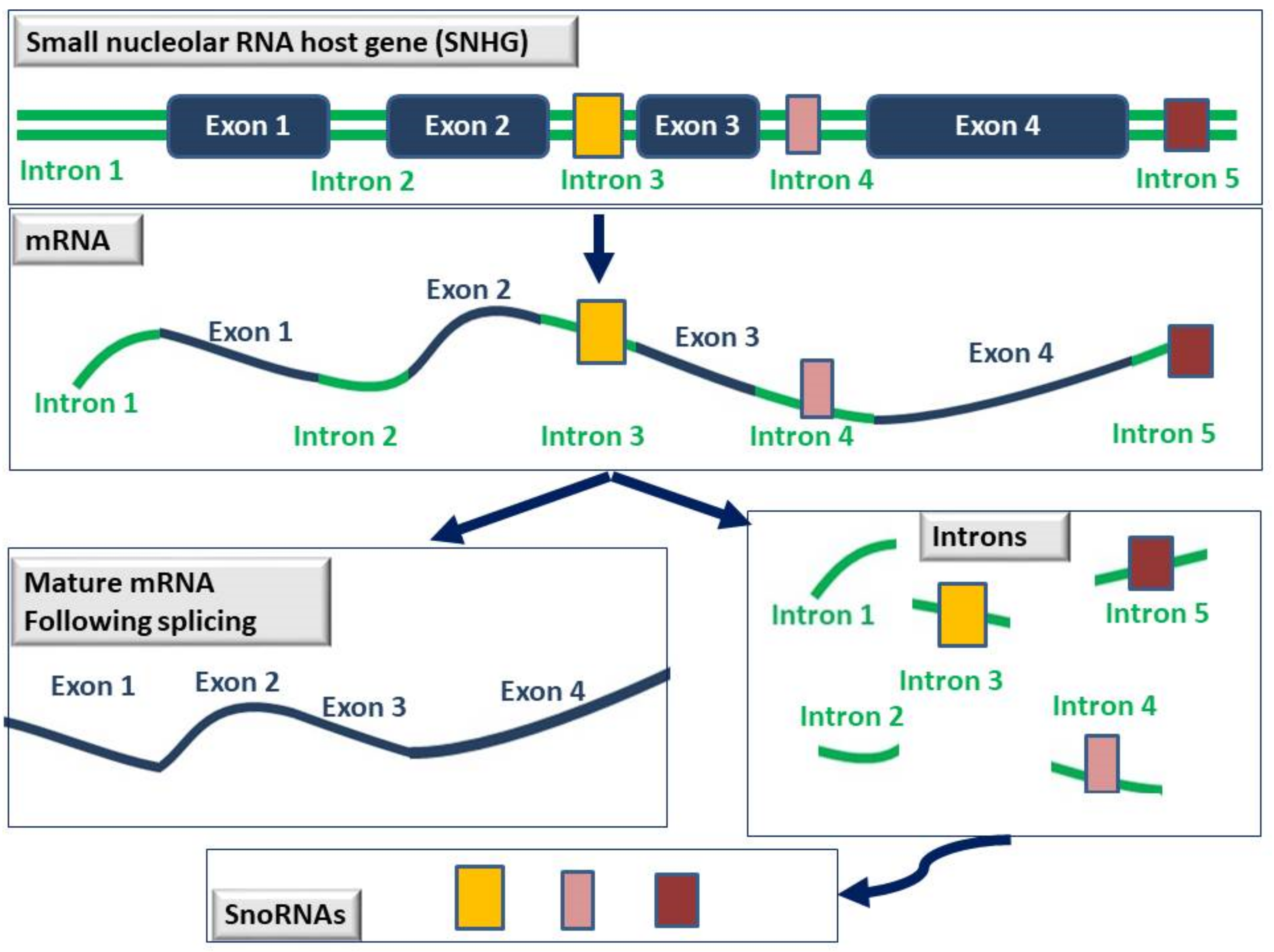
3. Small Nucleolar RNAs in Breast Cancer
References
- Xing, Z.; Zhang, M.; Liu, J.; Liu, G.; Feng, K.; Wang, X. LINC00337 induces tumor development and chemoresistance to paclitaxel of breast cancer by recruiting M2 tumor-associated macrophages. Mol. Immunol. 2021, 138, 1–9.
- Wang, H.; Yu, S.; Peng, H.; Shu, Y.; Zhang, W.; Zhu, Q.; Wu, Y.; Xu, Y.; Yan, J.; Xiang, H. Long noncoding RNA Linc00337 functions as an E2F1 co-activator and promotes cell proliferation in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 216.
- Zhang, X.; Gong, J.; Lu, J.; Chen, J.; Zhou, Y.; Li, T.; Ding, L. Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am. J. Transl. Res. 2019, 11, 6075–6083.
- Wei, B.; Kong, W.; Mou, X.; Wang, S. Comprehensive analysis of tumor immune infiltration associated with endogenous competitive RNA networks in lung adenocarcinoma. Pathol. Res. Pract. 2019, 215, 159–170.
- Hu, B.; Wang, X.; Li, L. Long noncoding RNA LINC00337 promote gastric cancer proliferation through repressing p21 mediated by EZH2. Am. J. Transl. Res. 2019, 11, 3238–3245.
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Busselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789.
- Qiu, S.Q.; Waaijer, S.J.H.; Zwager, M.C.; de Vries, E.G.E.; van der Vegt, B.; Schroder, C.P. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018, 70, 178–189.
- Kim, J.; Bae, J.S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat. Inflamm. 2016, 2016, 6058147.
- Ostuni, R.; Kratochvill, F.; Murray, P.J.; Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015, 36, 229–239.
- Sun, J.; Yang, J.; Lv, K.; Guan, J. Long non-coding RNA LINC00460 predicts poor survival and promotes cell viability in pancreatic cancer. Oncol. Lett. 2020, 20, 1369–1375.
- Hou, P.; Meng, S.; Li, M.; Lin, T.; Chu, S.; Li, Z.; Zheng, J.; Gu, Y.; Bai, J. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J. Exp. Clin. Cancer Res. 2021, 40, 52.
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Hajiesmaeili, M. Long intergenic non-protein coding RNA 460: Review of its role in carcinogenesis. Pathol. Res. Pract. 2021, 225, 153556.
- Hong, W.; Ying, H.; Lin, F.; Ding, R.; Wang, W.; Zhang, M. lncRNA LINC00460 Silencing Represses EMT in Colon Cancer through Downregulation of ANXA2 via Upregulating miR-433–3p. Mol. Ther. Nucleic Acids 2020, 19, 1209–1218.
- Zhu, Y.; Yang, L.; Chong, Q.Y.; Yan, H.; Zhang, W.; Qian, W.; Tan, S.; Wu, Z.; Lobie, P.E.; Zhu, T. Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR-489–5p/FGF7/AKT axis. Cancer Manag. Res. 2019, 11, 5983–6001.
- Dong, Y.; Quan, H.Y. Downregulated LINC00460 inhibits cell proliferation and promotes cell apoptosis in prostate cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6070–6078.
- Liu, X.; Wen, J.; Wang, H.; Wang, Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338–3p. Biomed. Pharmacother. 2018, 108, 1022–1028.
- Li, K.; Sun, D.; Gou, Q.; Ke, X.; Gong, Y.; Zuo, Y.; Zhou, J.K.; Guo, C.; Xia, Z.; Liu, L.; et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018, 420, 80–90.
- Liu, Y.; He, D.; Xiao, M.; Zhu, Y.; Zhou, J.; Cao, K. Long noncoding RNA LINC00518 induces radioresistance by regulating glycolysis through an miR-33a-3p/HIF-1alpha negative feedback loop in melanoma. Cell Death Dis. 2021, 12, 245.
- Chang, L.; Hu, Z.; Zhou, Z.; Zhang, H. Linc00518 Contributes to Multidrug Resistance Through Regulating the MiR-199a/MRP1 Axis in Breast Cancer. Cell Physiol. Biochem. 2018, 48, 16–28.
- Wang, H.B.; Wei, H.; Wang, J.S.; Li, L.; Chen, A.Y.; Li, Z.G. Down-regulated expression of LINC00518 prevents epithelial cell growth and metastasis in breast cancer through the inhibition of CDX2 methylation and the Wnt signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 708–723.
- Han, X.; Zhang, S. Role of Long Non-Coding RNA LINC00641 in Cancer. Front. Oncol 2021, 11, 829137.
- Zhang, Y.; Yu, R.; Li, L. LINC00641 hinders the progression of cervical cancer by targeting miR-378a-3p/CPEB3. J. Gene Med. 2020, 22, e3212.
- Yang, J.; Yu, D.; Liu, X.; Changyong, E.; Yu, S. LINC00641/miR-4262/NRGN axis confines cell proliferation in glioma. Cancer Biol. Ther. 2020, 21, 758–766.
- Mao, Q.; Lv, M.; Li, L.; Sun, Y.; Liu, S.; Shen, Y.; Liu, Z.; Luo, S. Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194–5p. J. Cell Physiol. 2020, 235, 2668–2675.
- Li, Y.; Zhao, L.; Zhao, P.; Liu, Z. Long non-coding RNA LINC00641 suppresses non-small-cell lung cancer by sponging miR-424–5p to upregulate PLSCR4. Cancer Biomark. 2019, 26, 79–91.
- Li, Z.; Hong, S.; Liu, Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197–3p/KLF10/PTEN/PI3K/AKT cascade. Biochem. Biophys. Res. Commun. 2018, 503, 1825–1829.
- Yang, F.; Xiao, Z.; Zhang, S. Knockdown of miR-194–5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/beta-catenin signaling pathway. Int. J. Mol. Med. 2018, 42, 3355–3363.
- Meng, D.F.; Shao, H.; Feng, C.B. LINC00894 Enhances the Progression of Breast Cancer by Sponging miR-429 to Regulate ZEB1 Expression. OncoTargets Ther. 2021, 14, 3395–3407.
- Ye, Z.; He, Q.; Wang, Q.; Lin, Y.; Cen, K.; Chen, X. LINC00922 promotes the proliferation, migration, invasion and EMT process of liver cancer cells by regulating miR-424–5p/ARK5. Mol. Cell. Biochem. 2021, 476, 3757–3769.
- Wang, Y.; Dong, T.; Wang, P.; Li, S.; Wu, G.; Zhou, J.; Wang, Z. LINC00922 regulates epithelial-mesenchymal transition, invasive and migratory capacities in breast cancer through promoting NKD2 methylation. Cell. Signal. 2021, 77, 109808.
- Wang, L.; Ren, C.; Xu, Y.; Yang, L.; Chen, Y.; Zhu, Y. The LINC00922 aggravates ovarian cancer progression via sponging miR-361–3p. J. Ovarian Res. 2021, 14, 77.
- Liang, T.; Wang, B.; Li, J.; Liu, Y. LINC00922 Accelerates the Proliferation, Migration and Invasion of Lung Cancer Via the miRNA-204/CXCR4 Axis. Med. Sci. Monit. 2019, 25, 5075–5086.
- Grunert, M.; Appelt, S.; Dunkel, I.; Berger, F.; Sperling, S.R. Altered microRNA and target gene expression related to Tetralogy of Fallot. Sci. Rep. 2019, 9, 19063.
- Arnold, K.M.; Pohlig, R.T.; Sims-Mourtada, J. Co-activation of Hedgehog and Wnt signaling pathways is associated with poor outcomes in triple negative breast cancer. Oncol. Lett. 2017, 14, 5285–5292.
- Li, Z.; Li, Y.; Wang, N.; Yang, L.; Zhao, W.; Zeng, X. miR-130b targets NKD2 and regulates the Wnt signaling to promote proliferation and inhibit apoptosis in osteosarcoma cells. Biochem. Biophys. Res. Commun. 2016, 471, 479–485.
- Cao, B.; Yang, W.; Jin, Y.; Zhang, M.; He, T.; Zhan, Q.; Herman, J.G.; Zhong, G.; Guo, M. Silencing NKD2 by Promoter Region Hypermethylation Promotes Esophageal Cancer Progression by Activating Wnt Signaling. J. Thorac. Oncol. 2016, 11, 1912–1926.
- Dong, Y.; Cao, B.; Zhang, M.; Han, W.; Herman, J.G.; Fuks, F.; Zhao, Y.; Guo, M. Epigenetic silencing of NKD2, a major component of Wnt signaling, promotes breast cancer growth. Oncotarget 2015, 6, 22126–22138.
- Tian, Y.H.; Jia, L.W.; Liu, Z.F.; Chen, Y.H. LINC01087 inhibits glioma cell proliferation and migration, and increases cell apoptosis via miR-384/Bcl-2 axis. Aging 2021, 13, 20808–20819.
- Naorem, L.D.; Prakash, V.S.; Muthaiyan, M.; Venkatesan, A. Comprehensive analysis of dysregulated lncRNAs and their competing endogenous RNA network in triple-negative breast cancer. Int. J. Biol. Macromol. 2020, 145, 429–436.
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66.
- Pareja, F.; Reis-Filho, J.S. Triple-negative breast cancers—A panoply of cancer types. Nat. Rev. Clin. Oncol. 2018, 15, 347–348.
- De Palma, F.D.E.; del Monaco, V.; Pol, J.G.; Kremer, M.; D’Argenio, V.; Stoll, G.; Montanaro, D.; Uszczynska-Ratajczak, B.; Klein, C.C.; Vlasova, A.; et al. The abundance of the long intergenic non-coding RNA 01087 differentiates between luminal and triple-negative breast cancers and predicts patient outcome. Pharmacol. Res. 2020, 161, 105249.
- Tu, Z.; Schmoellerl, J.; Mariani, O.; Zheng, Y.; Hu, Y.; Vincent-Salomon, A.; Karnoub, A.E. The LINC01119-SOCS5 axis as a critical theranostic in triple-negative breast cancer. NPJ Breast Cancer 2021, 7, 69.
- Han, Y.; Wang, X.; Mao, E.; Shen, B.; Huang, L. Analysis of Differentially Expressed lncRNAs and mRNAs for the Identification of Hypoxia-Regulated Angiogenic Genes in Colorectal Cancer by RNA-Seq. Med. Sci. Monit. 2019, 25, 2009–2015.
- Chandrashekaran, I.R.; Mohanty, B.; Linossi, E.M.; Dagley, L.F.; Leung, E.W.; Murphy, J.M.; Babon, J.J.; Nicholson, S.E.; Norton, R.S. Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5. Biochemistry 2015, 54, 4672–4682.
- Cooney, R.N. Suppressors of cytokine signaling (SOCS): Inhibitors of the JAK/STAT pathway. Shock 2002, 17, 83–90.
- Song, J.; Zhang, S.; Sun, Y.; Gu, J.; Ye, Z.; Sun, X.; Tang, Q. A Radioresponse-Related lncRNA Biomarker Signature for Risk Classification and Prognosis Prediction in Non-Small-Cell Lung Cancer. J. Oncol. 2021, 2021, 4338838.
- Li, Z.; Li, Y.; Wang, X.; Liang, Y.; Luo, D.; Han, D.; Li, C.; Chen, T.; Zhang, H.; Liu, Y.; et al. LINC01977 Promotes Breast Cancer Progression and Chemoresistance to Doxorubicin by Targeting miR-212–3p/GOLM1 Axis. Front. Oncol 2021, 11, 657094.
- Xu, R.; Ji, J.; Zhang, X.; Han, M.; Zhang, C.; Xu, Y.; Wei, Y.; Wang, S.; Huang, B.; Chen, A.; et al. PDGFA/PDGFRalpha-regulated GOLM1 promotes human glioma progression through activation of AKT. J. Exp. Clin. Cancer Res. 2017, 36, 193.
- Kladney, R.D.; Cui, X.; Bulla, G.A.; Brunt, E.M.; Fimmel, C.J. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology 2002, 35, 1431–1440.
- Tian, Y.; Zhou, J.; Zou, Y.; Luo, B.; Liu, Q.; Cao, X. Upregulated long noncoding RNAs LINC02163 and FEZF1-AS1 exert oncogenic roles in colorectal cancer. Anticancer Drugs 2021, 32, 66–73.
- Qin, C.; Jin, L.; Li, J.; Zha, W.; Ding, H.; Liu, X.; Zhu, X. Long Noncoding RNA LINC02163 Accelerates Malignant Tumor Behaviors in Breast Cancer by Regulating the MicroRNA-511–3p/HMGA2 Axis. Oncol. Res. 2020, 28, 483–495.
- Abdollahzadeh, R.; Daraei, A.; Mansoori, Y.; Sepahvand, M.; Amoli, M.M.; Tavakkoly-Bazzaz, J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J. Cell. Physiol. 2019, 234, 10080–10100.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Zaker, S.R.; Ghaedi, K. Downregulation of LINC02615 Is Correlated with The Breast Cancer Progress: A Novel Biomarker for Differential Identification of Breast Cancer Tissues. Cell J. 2021, 23, 414–420.
- Qin, Y.; Sun, W.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers. OncoTargets Ther. 2020, 13, 7699–7717.
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.; Thompson, I.; Yuan, Z.M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014, 33, 1348–1358.
- Jiang, H.; Li, X.; Wang, W.; Dong, H. Long non-coding RNA SNHG3 promotes breast cancer cell proliferation and metastasis by binding to microRNA-154–3p and activating the notch signaling pathway. BMC Cancer 2020, 20, 838.
- Pei, X.; Wang, X.; Li, H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int. J. Biol. Macromol. 2018, 118, 24–30.
- Zheng, S.; Li, M.; Miao, K.; Xu, H. SNHG1 contributes to proliferation and invasion by regulating miR-382 in breast cancer. Cancer Manag. Res. 2019, 11, 5589–5598.
- Zuo, Y.; Qu, C.; Tian, Y.; Wen, Y.; Xia, S.; Ma, M. The HIF-1/SNHG1/miR-199a-3p/TFAM axis explains tumor angiogenesis and metastasis under hypoxic conditions in breast cancer. Biofactors 2021, 47, 444–460.
- Kang, Y.; Wan, L.; Wang, Q.; Yin, Y.; Liu, J.; Liu, L.; Wu, H.; Zhang, L.; Zhang, X.; Xu, S.; et al. Long noncoding RNA SNHG1 promotes TERT expression by sponging miR-18b-5p in breast cancer. Cell Biosci. 2021, 11, 169.
- Wan, Q.; Tang, M.; Sun, S.L.; Hu, J.; Sun, Z.J.; Fang, Y.T.; He, T.C.; Zhang, Y. SNHG3 promotes migration, invasion, and epithelial-mesenchymal transition of breast cancer cells through the miR-186–5p/ZEB1 axis. Am. J. Transl Res. 2021, 13, 585–600.
- Li, Y.; Zhao, Z.; Liu, W.; Li, X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl. Biochem. Biotechnol. 2020, 191, 1084–1099.
- Ma, Q.; Qi, X.; Lin, X.; Li, L.; Chen, L.; Hu, W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum. Cell 2020, 33, 232–242.
- Chi, J.R.; Yu, Z.H.; Liu, B.W.; Zhang, D.; Ge, J.; Yu, Y.; Cao, X.C. SNHG5 Promotes Breast Cancer Proliferation by Sponging the miR-154–5p/PCNA Axis. Mol. Ther. Nucleic Acids 2019, 17, 138–149.
- Li, K.; Ma, Y.B.; Tian, Y.H.; Xu, X.L.; Gao, Y.; He, Y.Q.; Pan, W.T.; Zhang, J.W.; He, C.J.; Wei, L. Silencing lncRNA SNHG6 suppresses proliferation and invasion of breast cancer cells through miR-26a/VASP axis. Pathol. Res. Pract. 2019, 215, 152575.
- Gao, Y.T.; Zhou, Y.C. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6588–6595.
- Luo, X.; Song, Y.; Tang, L.; Sun, D.H.; Ji, D.G. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7788–7797.
- Sun, X.; Huang, T.; Liu, Z.; Sun, M.; Luo, S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur. J. Pharmacol. 2019, 856, 172407.
- Zhang, L.; Fu, Y.; Guo, H. c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer. J. Breast Cancer 2019, 22, 533–547.
- Li, Y.; Guo, X.; Wei, Y. LncRNA SNHG7 inhibits proliferation and invasion of breast cancer cells by regulating miR-15a expression. J. BUON 2020, 25, 1792–1798.
- Chen, Z.; Zhong, T.; Li, T.; Zhong, J.; Tang, Y.; Liu, Z.; Ling, B.; Wang, L. LncRNA SNHG15 modulates gastric cancer tumorigenesis by impairing miR-506–5p expression. Biosci. Rep. 2021, 41, BSR20204177.
- Liu, L.B.; Jiang, Z.J.; Jiang, X.L.; Wang, S. Up-regulation of SNHG15 facilitates cell proliferation, migration, invasion and suppresses cell apoptosis in breast cancer by regulating miR-411–5p/VASP axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1899–1912.
- Xie, S.D.; Qin, C.; Jin, L.D.; Wang, Q.C.; Shen, J.; Zhou, J.C.; Chen, Y.X.; Huang, A.H.; Zhao, W.H.; Wang, L.B. Long noncoding RNA SNHG14 promotes breast cancer cell proliferation and invasion via sponging miR-193a-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2461–2468.
- Kong, Q.; Qiu, M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211–3p. Biochem. Biophys. Res. Commun. 2018, 495, 1594–1600.
- Cai, C.; Huo, Q.; Wang, X.; Chen, B.; Yang, Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem. Biophys. Res. Commun. 2017, 485, 272–278.
- Du, S.M. The SNHG16/miR-30a axis promotes breast cancer cell proliferation and invasion by regulating RRM2. Neoplasma 2020, 67, 567–575.
- Du, Y.; Wei, N.; Hong, J.; Pan, W. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124–3p. Cancer Cell Int. 2020, 20, 40.
- Guan, Y.X.; Zhang, M.Z.; Chen, X.Z.; Zhang, Q.; Liu, S.Z.; Zhang, Y.L. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J. Cell Biochem. 2018, 119, 7971–7981.




