Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | José Pérez de la Lastra | -- | 3731 | 2022-09-28 10:15:52 | | | |
| 2 | Sirius Huang | Meta information modification | 3731 | 2022-09-29 03:49:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lastra, J.P.D.L.; Andrés, C.M.C.; Juan, C.A.; Plou, F.J.; Lebeña, E. Hydrogen Peroxide Formation and Elimination in Mammalian Cells. Encyclopedia. Available online: https://encyclopedia.pub/entry/27825 (accessed on 05 March 2026).
Lastra JPDL, Andrés CMC, Juan CA, Plou FJ, Lebeña E. Hydrogen Peroxide Formation and Elimination in Mammalian Cells. Encyclopedia. Available at: https://encyclopedia.pub/entry/27825. Accessed March 05, 2026.
Lastra, José Pérez De La, Celia María Curieses Andrés, Celia Andrés Juan, Francisco J Plou, Eduardo Lebeña. "Hydrogen Peroxide Formation and Elimination in Mammalian Cells" Encyclopedia, https://encyclopedia.pub/entry/27825 (accessed March 05, 2026).
Lastra, J.P.D.L., Andrés, C.M.C., Juan, C.A., Plou, F.J., & Lebeña, E. (2022, September 28). Hydrogen Peroxide Formation and Elimination in Mammalian Cells. In Encyclopedia. https://encyclopedia.pub/entry/27825
Lastra, José Pérez De La, et al. "Hydrogen Peroxide Formation and Elimination in Mammalian Cells." Encyclopedia. Web. 28 September, 2022.
Copy Citation
Hydrogen peroxide (H2O2) is a compound involved in some mammalian reactions and processes. It modulates and signals the redox metabolism of cells by acting as a messenger together with hydrogen sulfide (H2S) and the nitric oxide radical (•NO), activating specific oxidations that determine the metabolic response.
hydrogen peroxide H2O2
metabolic response
1. Introduction
Hydrogen peroxide H2O2, also known as dioxygenane or dioxygen, is a strongly oxidizing compound with a particularly unpleasant odour that decomposes into oxygen and water, releasing large amounts of heat. Although non-flammable, it is a strong oxidizer that can cause combustion when it comes into contact with organic material or metals such as copper, silver or bronze [1].
Because of its chemical properties, hydrogen peroxide is used in many areas of human activity, such as healthcare, as antiseptic, antimicrobial and antibacterial agent [2].
In mammals, in terms of redox signalling and regulation, H2O2 is an endogenous oxidant [3]. It is the most stable of the reactive oxygen species ROS, clearly involved in the regulation of protein function [4], and under certain circumstances is also a precursor of hydroxyl radical •OH and hypochlorous acid HOCl [5].
At micromolar concentrations, H2O2 is reactive, and at high concentrations it can damage energy-transforming cell systems, e.g., by inactivating the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase [6]. In the Fenton reaction, soluble Fe(II) donates an electron to an H2O2 molecule, causing it to decompose into hydroxyl radicals •OH + −OH, which react at diffusion rates and can randomly oxidise virtually any organic molecule. Hydroxyl radicals kill by DNA damage [7].
Until a few decades ago, H2O2 was considered an undesirable and harmful product for metabolism because it is a by-product of oxidative stress in cells [8]. Instead, it has come to the fore in recent years as a key redox signalling molecule in a variety of biological processes, including cell differentiation and proliferation, inflammation, tissue repair, circadian rhythm, and even ageing [9]. Because of its relative stability, with a cellular half-life of around 10−3 s, its diffusivity and its selective reactivity, H2O2 is often presented as the most important redox signal molecule [10].
H2O2 can act as a signalling molecule or, conversely, cause oxidative damage to biomolecules (a condition known as oxidative stress). This ambivalence depends on the cellular context, the local concentration of H2O2 and the kinetics of its production and elimination [11]. Under healthy steady-state conditions, their production and elimination are balanced, but under conditions of oxidative stress, superoxide anion •O2− and thus H2O2, are overproduced.
Hydrogen peroxide is formed as a product of the monovalent reduction of superoxide or by the divalent reduction of oxygen [12]. The main source of hydrogen peroxide is enzyme-catalysed superoxide dismutation, but it can also result from the two-electron reduction of oxygen in reactions catalysed by oxidases such as xanthine oxidase, glucose oxidase, amino acid oxidase, urate oxidase, and others. Since hydrogen peroxide is uncharged with a pKa≈10.8 at neutral pH, it can easily penetrate biological membranes [13]. Hydrogen peroxide is a strong oxidant, but due to its slow reaction kinetics with the biomolecules, it is relatively unreactive. Therefore, it can accumulate in cells and tissues at relatively high concentrations [14]. H2O2 is capable of directly oxidize many molecules and inactivating certain enzymes with thiol groups or methionine residues in their active site [15]. Hydrogen peroxide is a weak reducing agent, with an E°’ (•O2−, 2H+/H2O2) of +0.940 V, and an oxidizing agent with an E°’(H2O2, H+/H2O, HO−) of +0.320 V.
2. Hydrogen Peroxide Formation
It has been known for more than 60 years that hydrogen peroxide forms naturally in living organisms. H2O2 is not a free radical, but it is a reactive form of great importance because it can form the hydroxyl radical •OH in the presence of metals such as iron, in the known Fenton reaction. In mammalian cells, two forms of H2O2 production coexist, defined as enzymatic and non-enzymatic generation. In essence, the non-enzymatic one derives from the reduction by e− and H+ of the •O2− anion, which in turn comes from the reduction of O2 in the mitochondrial respiration pathway (complexes I to IV) and which takes place in the cellular mitochondrial matrix.
The enzymatic pathway starts from the •O2− anion, and involves the enzyme superoxide dismutase, SOD1, SOD2 and SOD3. These mechanisms are explained in detail in the following subsections.
2.1. Non-Enzymatic Generation of H2O2
ROS are also formed in the mitochondrial matrix by partial reduction of O2 to •O2− and subsequent reduction of the superoxide radical by e− and H+ to H2O2 and H2O. Until a few years ago, this “leakage” of electrons from the chain was considered an altered process, however, it has now been proposed that mitochondrial •O2− and H2O2 may be involved in redox reaction-dependent signalling processes [16], as well as in the biological clock of ageing [17], and even act as an indicator of the proper functioning of the electron transport chain ETC. Inside mitochondrial matrix, the harmful effects of ROS include structural changes on mitDNA and oxidation of lipids and proteins that perform numerous functions. It is therefore important to keep ROS levels at physiological values, preventing them from increasing and triggering oxidation of biological molecules.
In this route, the final product is H2O, so O2, •O2− anion and hydrogen peroxide, in the presence of e− and H+, will also be spontaneously reduced to the final H2O stage. The Gibbs free energy of this reduction process is, at each step, negative.
Electrons derived from mitochondrial metabolism flow through complexes I-IV of the ETC in the mitochondrial inner membrane. The energy gradient of this process is used to pump protons (H+) to the intermembrane space. Mitochondrial respiration is coupled to ATP synthesis via ADP phosphorylation. Protons introduced by ATP synthase are used to reduce molecular O2 to •O2−, H2O2 and finally H2O, Figure 1:

Figure 1. O2 reduction to •O2− anion, H2O2 and finally H2O.
2.2. Enzymatic Generation of H2O2
•O2− is also formed in other cell organelles such as the endoplasmic reticulum, peroxisomes, and cytosol [18] and H2O2 production involves the enzyme superoxide dismutase, SOD-1, -2 and -3, Figure 2. In fact, there are enzymatic reactions within the cell that result in the generation of •O2−.
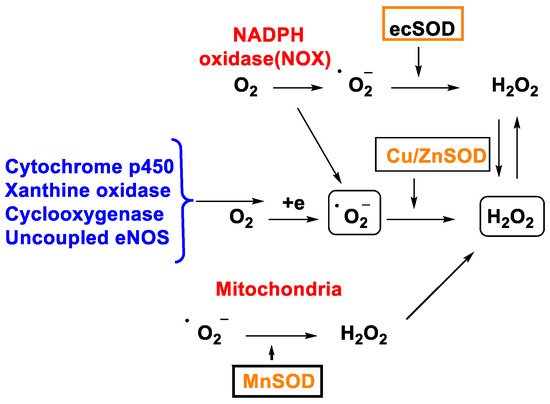
Figure 2. H2O2 production through different mechanisms.
In 1968, McCords and Fridovich discovered the enzyme superoxide dismutase SOD isolated from erythrocytes. SOD is the only enzyme capable of detoxifying the superoxide •O2− and is present at the mitochondrial level as well as in the cytoplasm and extracellular space [19]. There are three isoforms, SOD1 (Cu/Zn SOD) is the predominant •O2− scavenger and is localized in the cytoplasm, mitochondrial intermembrane space, nucleus, and lysosomes; SOD2 (Mn SOD) and SOD3 (Cu/Zn SOD) are localized in the mitochondrion and extracellular matrix respectively [20]. This group of metalloenzymes catalyses the dismutation of the superoxide radical to hydrogen peroxide and oxygen, Figure 3:
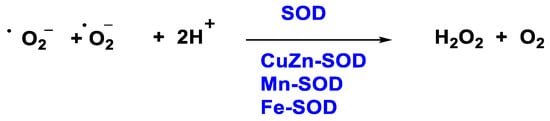
Figure 3. H2O2 production through SOD enzyme.
The SOD-catalysed dismutation of the superoxide radical can be represented as the following half-reactions, Figure 4:
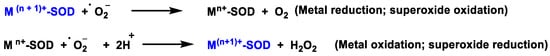
Figure 4. SOD-catalysed dismutation of the •O2−.
where M = Cu (n = 1); Mn (n = 2); Fe (n = 2). In this reaction the oxidation state of the metal cation ranges between n and n + 1.
In the catalytic cycle of cytosolic superoxide dismutase Cu/Zn SOD, which contains Cu and Zn in the catalytic site, SOD transforms the •O2− to O2 by reducing Cu(II) to Cu(I) in its active site. Then, another •O2− molecule causes the oxidation of Cu(I) to Cu(II), generating an H2O2 molecule at the end of the cycle. Zinc does not act in the catalytic cycle, it only helps to stabilize the enzyme, Figure 5, left. The catalytic cycle of mitochondrial superoxide dismutase Mn-SOD is similar, except for the use of Mn in the oxidation-reduction reactions, transiting between (Mn(III)) and (Mn(II)) [11], Figure 5, right.
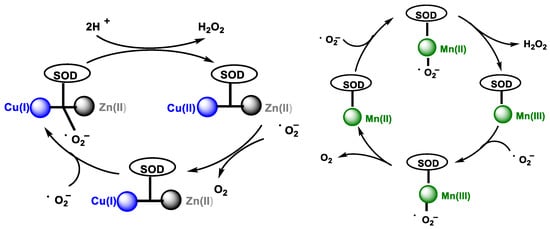
Figure 5. (Left): Cu/Zn SOD catalytic pathway. (Right): Mn SOD catalytic pathway.
NADPH oxidase is an enzyme that catalyses the reduction reaction of O2 to •O2− and/or H2O2 using NADPH as an electron donor [21]. It was first identified in phagocytic cells of the innate immune response and is involved in the “respiratory burst”, generating large amounts of •O2− to destroy invading pathogens.
In general, phase 1 enzymes can transform multiple substrates and catalyse different reactions. They are catalytic proteins of a very diverse nature including enzymes with monooxygenase activity, such as cytochrome P450s or flavin monooxygenase, various oxidases (alcohol dehydrogenase, aldehyde dehydrogenase, amino oxidases, aromatases), epoxide hydrolase, or hepatic and plasma esterases and amidases. Cytochrome P450s are undoubtedly the most prominent member of this group of enzymes and the one that has been most extensively studied [22].
Cytochrome P450s, responsible for detoxifying substances in the body, can carry out oxidation reactions that result in the generation of superoxide [23]. On the other hand, the enzyme xanthine oxidase catalyses the oxidation of hypoxanthine and xanthine to uric acid with the formation of hydrogen peroxide. Thus, the presence of enzymes whose activity leads to the formation of •O2− and H2O2 indicates that the production of these species is not a random event and that their production in the cell must therefore have a specific purpose beyond causing damage [14].
In humans, xanthine oxidase is normally found in the liver and not free in the blood [24]. The enzyme xanthine oxidase is a molybdoflavoenzyme, a flavo-protein one (FAD, as a cofactor) containing one molybdenum atom and four ferro-sulphur centres in its prosthetic group [25]. Mo is pentacoordinate. The main function of this metalloenzyme is to hydroxylate a number of substrates, such as hypoxanthine, which is converted to xanthine, which in turn is converted to uric acid. O2, which acts as an oxidant in both reactions, is converted to H2O2. At each oxidation step, xanthine oxidase XOR generates superoxide radical ion and hydrogen peroxide [26], Figure 6:
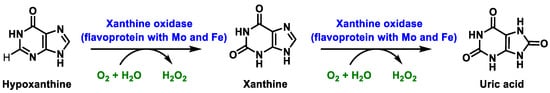
Figure 6. Xanthine oxidase XOR generates superoxide radical ion and hydrogen peroxide.
In mammals, this enzyme catalyses the hydroxylation of hypoxanthine to xanthine and xanthine to uric acid [27]. XOR uses hypoxanthine and oxygen (as an electron acceptor) to give rise to xanthine (eventually uric acid) and superoxide radical. In the active form of XOR, Mo forms two single bonds with the sulphur atom (thiol groups), two bonds with the oxygen atom (one with the oxo group and one with the hydroxyl group) and the fifth coordination position is occupied by a double bond with the sulphur atom. XOR converts xanthine to uric acid, Figure 7, the end product of the catabolism of purine bases in humans.
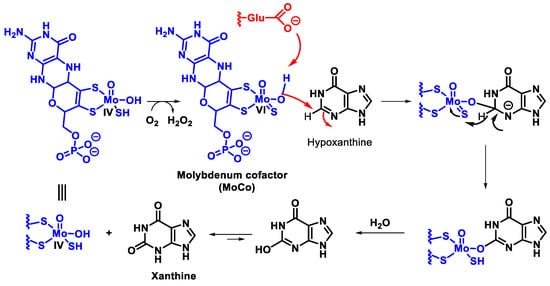
Figure 7. Mechanism of xanthine oxidase reaction.
The mechanism by which XOR converts xanthine to uric acid is not fully understood, but it has been suggested that a reduction and oxidation reaction occurs, as shown in the figure, resulting in oxidative damage to tissues [28], Figure 8.
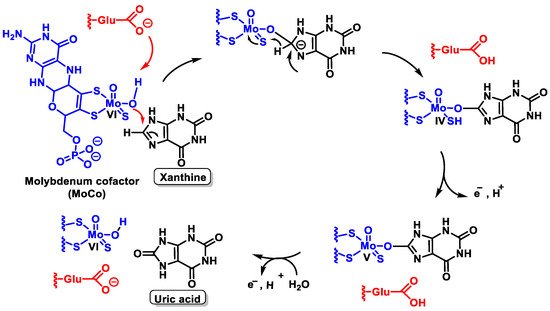
Figure 8. Possible mechanism by which XOR converts xanthine to uric acid.
Catalysis is initiated by base-assisted nucleophilic attack of the equatorial Mo-OH at the C-8 carbon of xanthine with hydrogen transfer from C-8 to MoS and moves from Mo=S to Mo-SH, which simultaneously results in the reduction of Mo(VI) to Mo(IV). Reoxidation of the molybdenum centre occurs by electron transfer to the other redox active sites of the enzyme, accompanied by deprotonation of the Mo- SH bond and cleavage of the bound product by hydroxide from the solvent to regenerate the Mo-OH group.
The peroxisome contains different proteins that can generate H2O2, such as urate oxidase, 1-α-hydroxyacid oxidase, and D-amino acid oxidase.
Peroxisomes are very common cytoplasmic organelles in the form of vesicles with a single membrane found in most eukaryotic cells. They are an oxidative organelle in which molecular oxygen serves as a co-substrate for the formation of hydrogen peroxide. Peroxisomes are named for their ability to produce hydrogen peroxide [29]. In addition, some microorganisms such as bacteria and mycoplasmas release hydrogen peroxide, which can cause damage to the host at the cellular level due to its ability to penetrate biological membranes.
Although urate oxidase is present in almost all living organisms, bacteria, fungi, plants, and animals, it is absent in many primates and in humans. In the human genome, there is a gene for urate oxidase that has been rendered non-functional by two mutations. Since urate oxidase is missing, uric acid is the end product of purine catabolism in humans [30][31].
Unlike other animals, humans do not have the enzyme urate oxidase [31] that converts uric acid to allantoin, a very soluble biomolecule, so they cannot break down excess uric acid, which is very poorly soluble. As a result, urate, which makes up most of the uric acid in the blood, is transported by plasma proteins such as albumin and alpha-globulins [32], Figure 9.
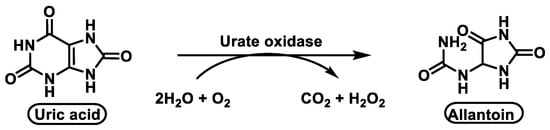
Figure 9. The enzyme urate oxidase converts uric acid into allantoin.
Previously, this metabolic pathway was thought to be carried out by a single enzyme, urate oxidase, but recent research has shown that two other enzymes are involved in this metabolic pathway, Figure 10. The metabolic pathway is initiated by urate oxidase, which produces the unstable 5-hydroxyisourate HIU, followed by hydrolysis by HIU hydrolase to form OHCU, which is also spontaneously degraded.
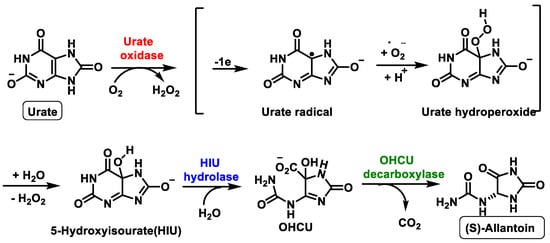
Figure 10. Schematic representation of the ureide pathway.
The third enzyme, OHCU decarboxylase, catalyses the de-carboxylation of OHCU, producing (S)-allantoin in a stereospecific manner [33], Figure 11.
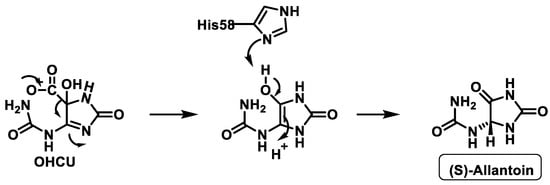
Figure 11. Proposed mechanism of OHCU decarboxylase.
The oxidoreductase enzyme oxidoreductin1 Ero1 catalyses the formation and isomerisation of protein disulphide bonds in the endoplasmic reticulum ER of eukaryotic cells. This luminal glycoprotein is tightly associated with the ER membrane and is essential for the oxidation of protein dithiols. Disulphide bond formation is an oxidative process and Ero1 is required for the introduction of oxidant equivalents into the ER and their direct transfer to the protein disulphide isomerase PDI. Ero1 is a major producer of H2O2 in the lumen of the ER endoplasmic reticulum [34]. Oxidative protein folding in the ER is an essential function of eukaryotic cells and requires the transmission of electrons between the different protein components. Ero1 reduces O2 to H2O2, its activity is allosterically and transcriptionally regulated by the response to unfolded ER proteins. The oxidative activity of Ero1 is linked to H2O2 production and consequently burdens cells with potentially toxic reactive oxygen species, so deregulated Ero1 activity impairs cell metabolism under certain conditions of oxidative ER stress [34].
3. Removal of Hydrogen Peroxide
In biological systems, H2O2 can be removed directly by the enzyme CAT, Figure 12, which uses H2O2 to generate molecular oxygen and water, whereas GPx uses H2O2 and reduced glutathione (GSH) to form water and oxidised glutathione GSSG. Fe(II) reacts with H2O2 in a Fenton reaction, producing •OH and OH−. Iron can also react with H2O2 to produce OOH− and H+ [35].
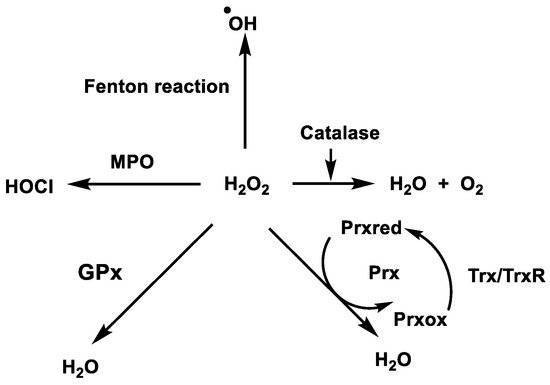
Figure 12. Enzymatic removal of hydrogen peroxide in biological systems.
While H2O2 is less reactive and more stable than •O2− [36], Figure 13, it generates hydroxyl radicals, one of the most reactive oxygen species known. Therefore, the removal of peroxide is of utmost importance to avoid oxidative damage.
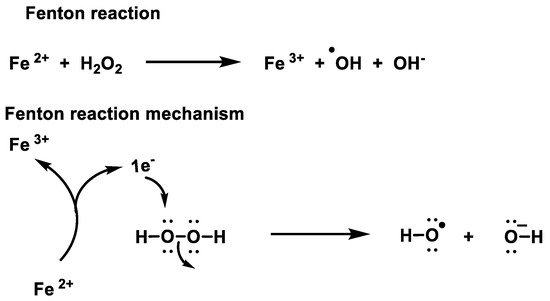
Figure 13. Mechanism of the Fenton reaction.
Non-enzymatic removal of H2O2 occurs via the Fenton reaction and enzymatic removal is carried out by catalases, glutathione peroxidases and peroxiredoxins.
3.1. Catalase
Catalase is an antioxidant enzyme found in most aerobic organisms. It catalyses the dismutation of H2O2 into water and oxygen. Most of these enzymes are homotetramers with a heme group on each subunit. In the catalase reaction, a two-electron transfer occurs between two hydrogen peroxide molecules, one acting as an electron donor and the other as an electron acceptor. The reaction mechanism proceeds in two steps. In the first step, catalase is oxidised by a peroxide molecule to form an intermediate called compound I. Compound I is characterised by a ferroxyl group containing FeIV and a porphyrin cation radical. In this reaction, a water molecule is formed (reaction 1). In the second step of the reaction, compound I is reduced by another peroxide molecule, returning the catalase to its initial state and producing water and dioxygen (reaction 2), Figure 14.
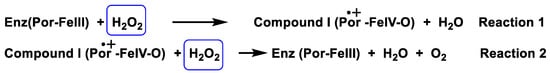
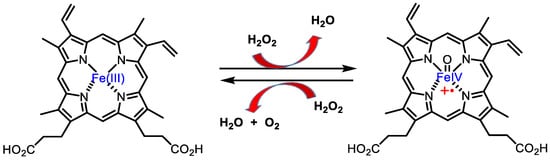
Figure 14. Catalase mechanism of hydrogen peroxide dismutation to water and oxygen.
Catalase is mainly localized to peroxisomes by the Pex5-mediated import pathway [37], as well as in the mitochondrial matrix of some metabolically highly active tissues, such as the heart and liver [38].
3.2. Glutathione Peroxidases GPx
It is in the cytosol and mitochondria. Its importance lies in the fact that it is the main antioxidant system at low levels of oxidative stress [39]. The term glutathione peroxidase GPx is associated with a family of multiple isoenzymes GPx1–8 that catalyse the reduction of H2O2 to water using glutathione as an electron donor [40][41]. The different isoforms can be divided into Se-dependent and Se-independent isoforms. The Se-independent form, also known as glutathione S-transferase GST, catalyses the detoxification of various xenobiotics, with the Se atom not involved in the catalytic reaction. The Se-dependent isoform, also referred to as Se-dependent GPx, consists of four subunits, and each subunit contains a Se atom in the active site bound to the amino acid cysteine. Except for mammalian GPx 5 and 6, all glutathione peroxidases are selenoproteins and contain a SeCys residue instead of Cys as part of their active site.
The highest GPx activity is detected in the liver, while medium activity is observed in the heart and lungs [42]. GPx enzyme cooperates with reduced glutathione GSH and is present in cells at high concentrations (millimoles). The GPx enzyme degrades peroxides to water or alcohol, while GSH is oxidised, reducing glutathione Se from the catalytic centre of the enzyme, Figure 15. Selenol (−SeH) in GPx, which contains selenocysteine Sec, reacts as selenolate with H2O2 to form selenic acid −SeOH, which is reduced by two molecules of GSH back to −SeH, forming GSSG and water [43]:
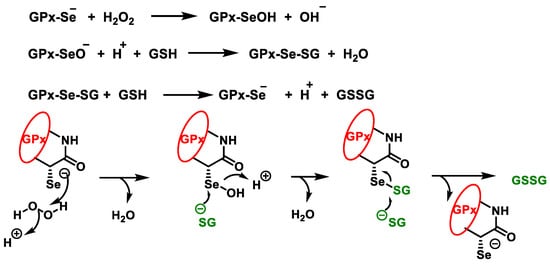
Figure 15. Catalytic cycle of glutathione peroxidase for the reduction of H2O2.
In the peroxidative part of the cycle, the selenol in GPx is oxidised by hydrogen peroxide to seleninic acid. The first GSH molecule forms a selenium disulphide with the seleninic acid, with the oxygen leaving as a water molecule. The second GSH molecule reduces the selenium disulphide by thiol-disulphide exchange, the GSSG is released, and the enzyme is regenerated to its selenol form to begin a new cycle.
The GSSG produced by the GPx enzyme in this process, as well as in other metabolic processes, must be constantly recycled in the cell for the peroxidative system to function properly. This is done by glutathione reductase GR, which is responsible for reducing GSSG to GSH. This disulphide bridge is reduced using NADPH as an electron source, Figure 16.
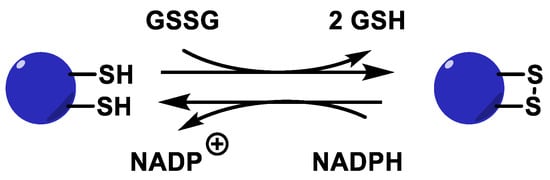
Figure 16. Glutathione reductase catalytic cycle.
3.3. Peroxiredoxins (Prx) and Thioredoxins (Trx)
Prx are a large family of antioxidant enzymes capable of breaking down peroxides that have cysteine in their active center [44]. These enzymes are responsible for controlling peroxide levels and are found in the cytosol, nucleus, membranes, mitochondria, Golgi complex, peroxisomes, and extracellular fluids. Prxs form a family of enzymes that, depending on the isoform or species, can detoxify hydrogen peroxides, peroxides of long-chain organic compounds, phospholipids and fatty acids, and peroxynitrite [45].
In the case of peroxiredoxin containing a cysteine (1-Cys-Prx or Prx6 isoform), the SOH formed during peroxide reduction is not attacked by the cysteine enzyme but is reduced using glutathione GSH as an electron source and forming glutathione disulphide GSSG, a mechanism like that of glutathione peroxidase. Reduction of SOH (1-Cys-Prx) or S-S (typical and atypical 2-Cys-Prx) allows regeneration of the active forms of Prx, Figure 17, left. Except for the Prx6 isoform, these enzymes contain two cysteines in the active site. One is a thiol that is oxidised to sulphenic acid SOH during the catalytic cycle. The other is a resolving cysteine that attacks SOH by forming an intermolecular S-S (typical 2-Cys-Prx or Prx1–Prx4 isoforms) or intramolecular (atypical 2-Cys-Prx or Prx5 isoform) disulphide bridge. The disulphide bridge formed in the catalytic cycle is reduced using reduced thioredoxin Trxred as an electron source to generate oxidised thioredoxin Trxox, Figure 17, right.
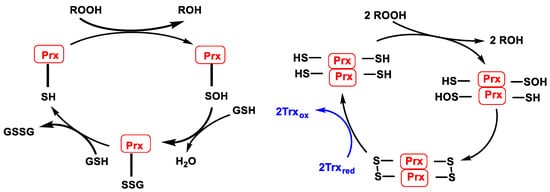
Figure 17. (Left): Catalytic cycle of Prx containing 1 catalytic cysteine (1-Cys-Prx). (Right): Catalytic cycle of typical Prx (2-Cys-Prx).
Atypical peroxiredoxins (atypical 2-Cys-Prx) are monomers with two cysteines participating in the catalytic cycle in a single subunit. In all cases, one cysteine SH is oxidised to sulfenic acid SOH by reducing peroxides ROOH. In the atypical 2-Cys-Prx, SOH is reduced by a cysteine present in the same subunit, resulting in an intramolecular disulphide bond that is also reduced by Trxred, Figure 18.
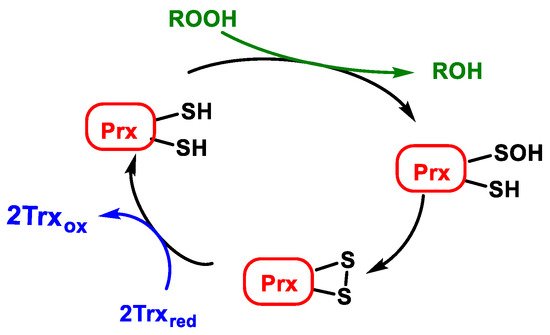
Figure 18. Catalytic cycle of atypical peroxiredoxins (atypical 2-Cys-Prx).
Trx are proteins that act as antioxidants by facilitating the reduction of other proteins through thiol-disulphide exchange on cysteine. Thioredoxin reductase TrxR is responsible for the reduction of Trxox to Trxred and requires NADPH as an electron donor source [46], Figure 19.
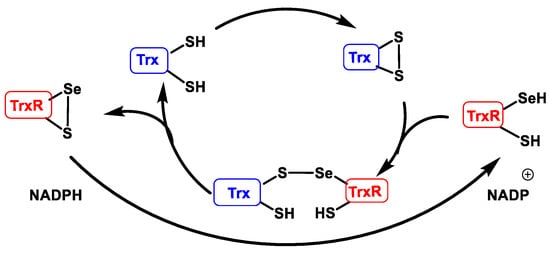
Figure 19. Thioredoxin reductase catalytic cycle (TrxR).
When used as an electron source in the catalytic peroxiredoxin cycle, thioredoxin Trx meets the thiol group SH of its two cysteines in disulphide form S-S. TrxR, which has a selenocysteine in selenol form SeSH and a SH at its catalytic site, performs the S-S reduction of Trx, forming an intermolecular sulphur-selenium bridge S-Se. This bridge is then reduced, creating an intramolecular S-Se bridge in TrxR and releasing the Trx in reduced form. TrxR regenerates into its reduced form using NAPDH as an electron source [47].
3.4. Hypochlorous Acid Formation (HOCl)
HOCl biologically falls into a group of small molecules known as reactive species RS synthesised by cells of the immune system (neutrophils and macrophages). HOCl plays a dual role, performing a bactericidal function against infections and, on the other hand, it can cause damage to the molecular structures and cells of the host organism. HOCl is a powerful antimicrobial oxidant, capable of modifying DNA, lipids and lipoproteins, reacting rapidly with the sulphur atom present in thiols and thioethers (cysteine and methionine) [48]. Excessive generation of HOCl can cause tissue damage and is thought to be important in the progression of a number of diseases including atherosclerosis, chronic inflammation and some cancers [49].
Myeloperoxidase is the most abundant protein in neutrophils and is the only peroxidase that catalyses the conversion of hydrogen peroxide and chloride to hypochlorous acid [50], Figure 20.
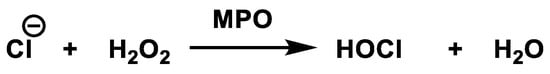
Figure 20. Conversion of hydrogen peroxide and chloride to hypochlorous acid.
When the human body is invaded by bacteria or viruses, our immune system responds immediately by sending an increased number of white blood cells called neutrophils to the site of invasion. Once activated, neutrophils produce through the enzyme myeloperoxidase large quantities of an oxidising solution (HOCl), which is highly effective in killing all pathogenic invading microbes.
References
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, e04691.
- Thomas, E.L.; Milligan, T.W.; Joyner, R.E.; Jefferson, M.M. Antibacterial activity of hydrogen peroxide and the lactoperoxidase-hydrogen peroxide-thiocyanate system against oral streptococci. Infect. Immun. 1994, 62, 529–535.
- Sies, H. Role of Metabolic H2O2 Generation: Redox Signaling And Oxidative Stress. J. Biol. Chem. 2014, 289, 8735–8741.
- Dickinson, B.C.; Chang, C.J. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011, 7, 504–511.
- Castagna, R.; Eiserich, J.P.; Budamagunta, M.S.; Stipa, P.; Cross, C.E.; Proietti, E.; Voss, J.C.; Greci, L. Hydroxyl radical from the reaction between hypochlorite and hydrogen peroxide. Atmos. Environ. 2008, 42, 6551–6554.
- Hyslop, P.A.; Chaney, M.O. Mechanism of GAPDH Redox Signaling by H2O2 Activation of a Two−Cysteine Switch. Int. J. Mol. Sci. 2022, 23, 4604.
- Mahaseth, T.; Kuzminov, A. Potentiation of hydrogen peroxide toxicity: From catalase inhibition to stable DNA-iron complexes. Mutat. Res. Rev. Mutat. Res. 2017, 773, 274–281.
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen peroxide–production, fate and role in redox signaling of tumor cells. Cell Commun. Signal. 2015, 13, 39.
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619.
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824.
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine, 4th ed.; Oxford University Press: Oxford, UK, 2015.
- Messner, K.R.; Imlay, J.A. Mechanism of Superoxide and Hydrogen Peroxide Formation by Fumarate Reductase, Succinate Dehydrogenase, and Aspartate Oxidase. J. Biol. Chem. 2002, 277, 42563–42571.
- Jiménez, M.; Lissarrague, M.S.; Bechthold, P.; González, E.; Jasen, P.; Juan, A. Ethanol adsorption on Ni doped Mo2C (001): A theoretical study. Top. Catal. 2022, 104, 839–847.
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496.
- Day, A.M.; Brown, J.D.; Taylor, S.R.; Rand, J.D.; Morgan, B.A.; Veal, E.A. Inactivation of a Peroxiredoxin by Hydrogen Peroxide Is Critical for Thioredoxin-Mediated Repair of Oxidized Proteins and Cell Survival. Mol. Cell 2012, 45, 398–408.
- Dada, L.A.; Chandel, N.S.; Ridge, K.M.; Pedemonte, C.; Bertorello, A.M.; Sznajder, J.I. Hypoxia-induced endocytosis of Na,K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-ζ. J. Clin. Investig. 2003, 111, 1057–1064.
- Nemoto, S.; Fergusson, M.M.; Finkel, T. SIRT1 Functionally Interacts with the Metabolic Regulator and Transcriptional Coactivator PGC-1α. J. Biol. Chem. 2005, 280, 16456–16460.
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxid. Med. Cell. Longev. 2019, 2019, 6175804.
- Fridovich, I. The Biology of Oxygen Radicals. Science 1978, 201, 875–880.
- Trachootham, D.; Lu, W.; Ogasawara, M.A.; Valle, N.R.-D.; Huang, P. Redox Regulation of Cell Survival. Antioxid. Redox Signal. 2008, 10, 1343–1374.
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015, 12, 5–23.
- Guengerich, F.P.; Munro, A.W. Unusual Cytochrome P450 Enzymes and Reactions. J. Biol. Chem. 2013, 288, 17065–17073.
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141.
- Ren, J.; Sowers, J.R.; Zhang, Y. Autophagy and Cardiometabolic Diseases: From Molecular Mechanisms to Translational Medicine; Elsevier: Amsterdam, The Netherlands, 2018.
- Nriagu, J.O. Encyclopedia of Environmental Health; Elsevier: Amsterdam, The Netherlands, 2011.
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine Oxidoreductase-Derived Reactive Species: Physiological and Pathological Effects. Oxid. Med. Cell. Longev. 2016, 2016, 3527579.
- Hille, R. Xanthine oxidoreductase. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–10.
- Becker, B.F. Towards the physiological function of uric acid. Free Radic. Biol. Med. 1993, 14, 615–631.
- Cooper, G. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000.
- Wu, X.W.; Lee, C.C.; Muzny, D.M.; Caskey, C.T. Urate oxidase: Primary structure and evolutionary implications. Proc. Natl. Acad. Sci. USA 1989, 86, 9412–9416.
- Kratzer, J.T.; Lanaspa, M.A.; Murphy, M.N.; Cicerchi, C.; Graves, C.L.; Tipton, P.A.; Ortlund, E.A.; Johnson, R.J.; Gaucher, E.A. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc. Natl. Acad. Sci. USA 2014, 111, 3763–3768.
- Liu-Bryan, R.; Terkeltaub, R. Tophus Biology and Pathogenesis of Monosodium Urate Crystal–Induced Inflammation. In Gout & Other Crystal Arthropathies; Terkeltaub, R., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2012; pp. 59–71.
- Kim, K.; Park, J.; Rhee, S. Structural and Functional Basis for (S)-Allantoin Formation in the Ureide Pathway. J. Biol. Chem. 2007, 282, 23457–23464.
- Zito, E. ERO1: A protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 2015, 83, 299–304.
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82, 969–974.
- Di Giorgio, F.P.; Carrasco, M.A.; Siao, M.C.; Maniatis, T.; Eggan, K. Non–cell autonomous effect of glia on motor neurons in an embryonic stem cell–based ALS model. Nat. Neurosci. 2007, 10, 608–614.
- Otera, H.; Fujiki, Y. Pex5p imports folded tetrameric catalase by interaction with Pex13p. Traffic 2012, 13, 1364–1377.
- Okumoto, K.; El Shermely, M.; Natsui, M.; Kosako, H.; Natsuyama, R.; Marutani, T.; Fujiki, Y. The peroxisome counteracts oxidative stresses by suppressing catalase import via Pex14 phosphorylation. eLife 2020, 9, e55896.
- Benner, P.; Hooper-Kyriakidis, P.; Stannard, D. Clinical Wisdom and Interventions in Acute and Critical Care; Springer: New York, NY, USA, 2011.
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970.
- Sena, L.A.; Chandel, N.S. Physiological Roles of Mitochondrial Reactive Oxygen Species. Mol. Cell 2012, 48, 158–167.
- Ďuračková, Z. Free Radicals and Antioxidants for Non-Experts. In Systems Biology of Free Radicals and Antioxidants; Laher, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–38.
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta BBA Gen. Subj. 2013, 1830, 3289–3303.
- Rhee, S.G.; Chae, H.Z.; Kim, K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005, 38, 1543–1552.
- Hall, A.; Nelson, K.; Poole, L.B.; Karplus, P.A. Structure-based Insights into the Catalytic Power and Conformational Dexterity of Peroxiredoxins. Antioxid. Redox Signal. 2010, 15, 795–815.
- Nordberg, J.; Arnér, E.S.J. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Review based on “Thioredoxin reductase—Interactions with the redox active compounds 1-chloro-2,4-dinitrobenzene and lipoic acid”. Free Radic. Biol. Med. 2001, 31, 1287–1312.
- Berndt, C.; Lillig, C.H.; Holmgren, A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: Implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1227–H1236.
- Tvrdá, E.; Benko, F. Chapter 1—Free radicals: What they are and what they do. In Pathology; Preedy, V.R., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 3–13.
- Casciaro, M.; Di Salvo, E.; Pace, E.; Ventura-Spagnolo, E.; Navarra, M.; Gangemi, S. Chlorinative stress in age-related diseases: A literature review. Immun. Ageing 2017, 14, 21.
- Davies, M.J. Myeloperoxidase-derived oxidation: Mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 2010, 48, 8–19.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.3K
Revisions:
2 times
(View History)
Update Date:
29 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





