Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xiaowei Xu | -- | 1289 | 2022-09-22 12:51:36 | | | |
| 2 | Dean Liu | Meta information modification | 1289 | 2022-09-23 04:57:07 | | | | |
| 3 | Dean Liu | -27 word(s) | 1262 | 2022-09-26 08:01:24 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yao, T.; Xiao, H.; Wang, H.; Xu, X. Proteolysis-targeting Chimeras for Drug Targeted Protein Research. Encyclopedia. Available online: https://encyclopedia.pub/entry/27480 (accessed on 01 March 2026).
Yao T, Xiao H, Wang H, Xu X. Proteolysis-targeting Chimeras for Drug Targeted Protein Research. Encyclopedia. Available at: https://encyclopedia.pub/entry/27480. Accessed March 01, 2026.
Yao, Tingting, Heng Xiao, Hong Wang, Xiaowei Xu. "Proteolysis-targeting Chimeras for Drug Targeted Protein Research" Encyclopedia, https://encyclopedia.pub/entry/27480 (accessed March 01, 2026).
Yao, T., Xiao, H., Wang, H., & Xu, X. (2022, September 22). Proteolysis-targeting Chimeras for Drug Targeted Protein Research. In Encyclopedia. https://encyclopedia.pub/entry/27480
Yao, Tingting, et al. "Proteolysis-targeting Chimeras for Drug Targeted Protein Research." Encyclopedia. Web. 22 September, 2022.
Copy Citation
Proteolysis-targeting chimera (PROTAC) is a heterobifunctional molecule. Typically, PROTAC consists of two terminals which are the ligand of the protein of interest (POI) and the specific ligand of E3 ubiquitin ligase, respectively, via a suitable linker. PROTAC degradation of the target protein is performed through the ubiquitin–proteasome system (UPS).
PROTAC
target protein
protein degradation
1. Introduction
Targeted protein degradation (TPD) is an emerging therapeutic modality that has the potential to solve the dilemma faced by traditional small molecule targeted therapy. Targeted protein degradation currently mainly degrades target proteins through ubiquitin–proteasome and lysosome. At present, molecular glue and PROTAC technology are the fastest growing in the field of targeted protein degradation [1].
Crews et al. introduced PROTAC for the first time in 2001, and PROTAC works by reducing protein levels rather than inhibiting protein function [2][3][4]. As a bifunctional small molecule compound, typically PROTAC consists of two terminals which are the ligand of the target protein and the specific ligand of E3 ubiquitin ligase, respectively, via a suitable linker [5][6][7][8][9][10]. PROTAC degrades target proteins through the ubiquitin–proteasome system (UPS). The general process is that PROTAC binds the target protein (POI) and E3 ligase to form a ternary complex, marking the target protein with the label of ubiquitination. The ubiquitinated proteins are recognized and degraded by the intracellular 26S proteasome [11][12][13][14][15][16][17][18][19] (Figure 1). The E3 ubiquitin ligase has approximately more than 600 members and is the most diverse component of the ubiquitin–proteasome system. The E3 ligases reported in the literature currently used in PROTAC mainly include Cereblon E3 ubiquitin ligase complex (CRBN), Von Hippel–Lindau-containing complex (VHL), inhibitor of apoptosis protein (IAP), and mouse double minute 2 (MDM2). The E3 ligases with the best effect and the highest frequency are mainly CRBN and VHL. Among them, the ligands of CRBN are mainly lenalidomide (Figure 2), thalidomide (Figure 2), and their analogs, while the ligands of VHL are mainly VHL-L (Figure 2) and 3-fluoro-VHL ligand [20][21][22][23][24] (Figure 2). PROTAC structurally connects two ligands through the linkers. The composition and length of the linker play an important role in PROTAC. Generally speaking, the composition and length of the linkers have different effects on degradation activity according to different targets. In addition, linked sites of the linkers also affect degradation activity. The binding sites of POI ligands and E3 ligase ligands are generally in the regions where the ligands are exposed to solvents. The connecting sites are generally connected by amide bonds, carbon atoms, or heteroatoms [25].
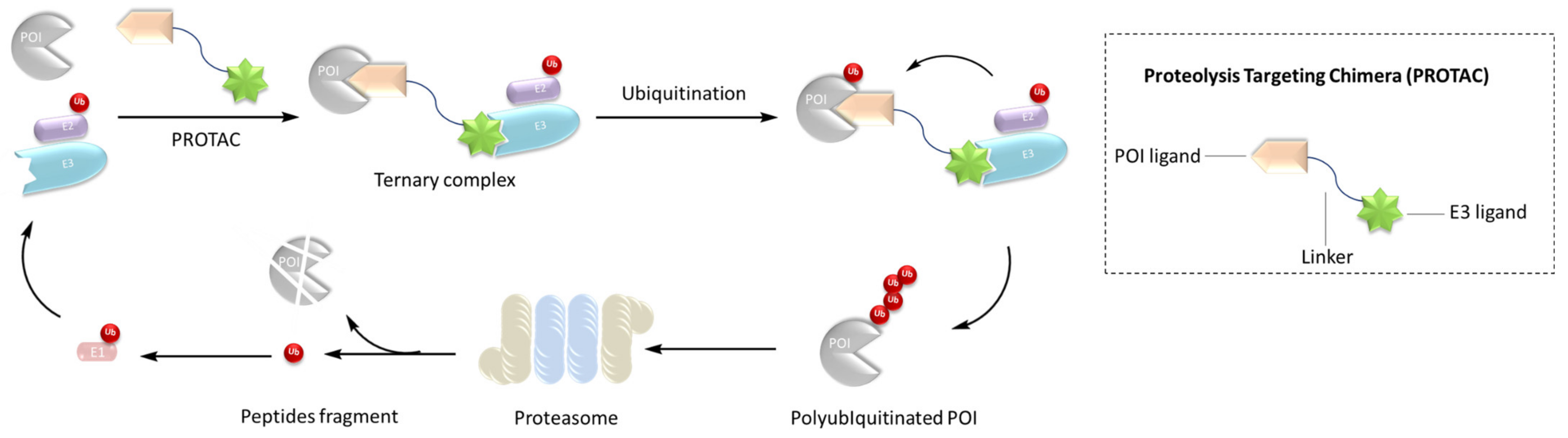
Figure 1. Mechanism of PROTAC.
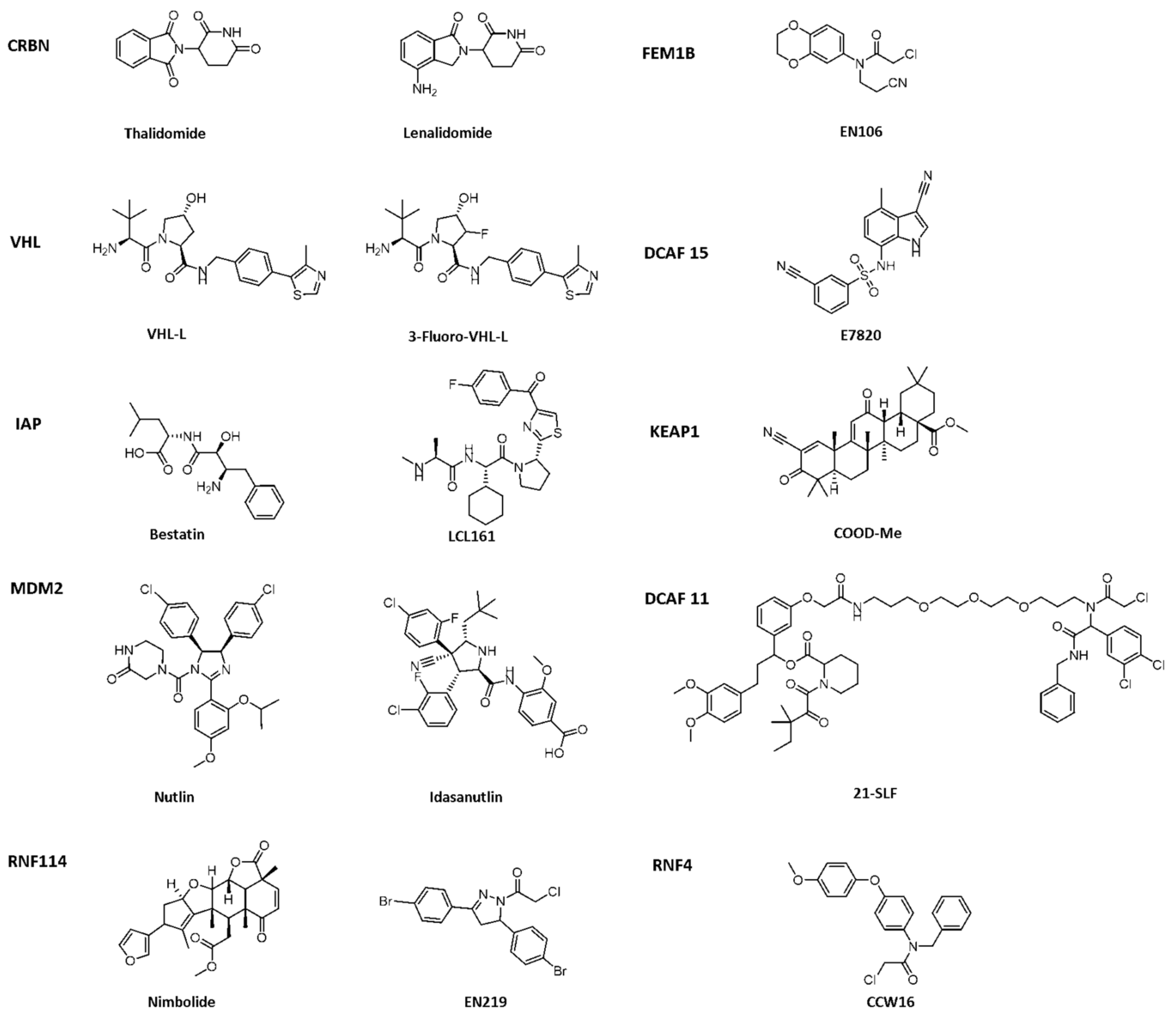
Figure 2. Existing E3 ligands used for targeted protein degradation applications.
Compared with traditional therapies, PROTAC technology has advantages such as wider scope of action, higher activity, and targeting “undruggable” targets. First, PROTAC can degrade the entire target protein to affect protein function, which is expected to solve the potential drug resistance problem faced by current traditional therapies; second, in theory, PROTAC can grasp the target protein through any corner and gap; therefore, PROTAC can target “undruggable target“; third, PROTAC can also affect non-enzymatic functions and expand the drug space of the target. So far, PROTACs have been successfully used to degrade several distinct target proteins associated with all kinds of illnesses, such as cancer, immune disorders, neurodegenerative conditions, cardiovascular diseases as well as viral infections [26][27][28]. In particular, 60 successful cases have demonstrated the effectiveness of PROTACs in degrading target proteins, two of which are currently in clinical trials for prostate and breast cancer treatment [29][30]. PROTAC has emerged as a fresh approach to medication development, providing a fresh method of treating disease.
2. Application of PROTAC in Anticancer
Cancer is one of the worst illnesses threatening human health. In recent years, the treatment of cancer is no longer confined to traditional surgery and radiotherapy and chemotherapy. Targeted therapy and immunotherapy play an important role in anti-cancer treatment. However, there are still no effective targeted drugs for “undruggable targets” such as KRAS and TP53 [31]. The ability to shift the target from “no drug” to “drug” is the most main benefit of PROTAC technology. Traditional targeted drugs need to be firmly bound to the target protein. Since PROTAC protein degrading agent can specifically “label” the target protein only by weakly binding with it, PROTAC degradation agent may solve about 80% of the current “undruggable” proteome. It is a timely help for patients who cannot carry out traditional targeted therapy [32].
3. Application of PROTAC in Immune Diseases
Immune inflammatory diseases are very common diseases in life, such as rheumatoid arthritis, systemic lupus erythematosus, ulcerative colitis, and so on. These diseases are threatening people’s health all the time. Scientists’ research on the treatment strategies for these diseases has never stopped. As an emerging strategy, PROTAC has penetrated into the treatment field of immune-inflammatory diseases. The following introduces the use of PROTAC in several applications in immune inflammatory disease targets.
4. Application of PROTAC in Neurodegenerative Diseases
Neurodegenerative diseases are an area in need of new therapies and molecular insights. The aggregation of misfolded proteins such as tau protein and α-synuclein protein is the main cause of such diseases, and they cannot be modulated by traditional small molecule drugs; therefore, the treatment of neurodegenerative diseases has always been a challenge. In recent years, the use of PROTAC technology to degrade target proteins has become a new treatment method. Therefore, PROTAC technology is expected to play a potential role in neurodegenerative diseases caused by protein aggregation.
5. Application of PROTAC in Cardiovascular Diseases
3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) catalyzes 3-hydroxy-3-methylglutaryl coenzyme A in the cholesterol synthesis pathway. HMGCR is a target of statins for the prevention and treatment of cardiovascular diseases [33][34]. In 2020, Luo’s team reported a series of PROTAC molecules [35], among which PROTAC 67 (Table 1) has the greatest impact on the HMGCR protein’s ability to degrade in Chinese hamster ovary SRD15 cells. Additionally, PROTAC 67 activates the sterol regulatory element-binding protein pathway (SREBP) and blocks cholesterol synthesis. That same year, Xiang’s group [36] reported two kinds of lovastatin acid and VHL ligand-conjugated HMGCR targeting PROTAC 68 (Table 1) and 69 (Table 1), and PROTAC 68 could effectively degrade HMGCR in HepG2 cells (DC50 = 120 nM). In vivo studies have shown that PROTAC 69 induces HMGCR breakdown and cholesterol reduction in mice with diet-induced hypercholesterolemia.
Table 1. Representative PROTACs for cardiovascular diseases.
| Protac | Target | Structure | Activity | Ref. | |
|---|---|---|---|---|---|
| DC50 | Dmax% | ||||
| 67 | HMGCR | 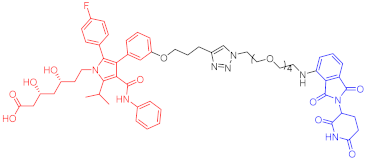 |
0.1 μM | - | [35] |
| 68 | 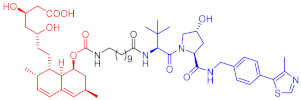 |
120 nM | 76 | [36] | |
| 69 | 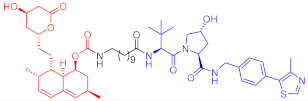 |
- | 56 | [36] | |
Note: red: molecule to bind to POI, black: linker, blue: ligand of E3 ligase.
6. Application of PROTAC in Antiviral
Infection with hepatitis C virus (HCV) is the main cause of chronic liver disease, in which the hepatitis C virus (HCV) NS3 protein plays an important role [37][38]. Although VX-950, an inhibitor of the NS3/4A protease, has been authorized for the treatment of HCV, patients are prone to develop drug resistance, so a new treatment method is urgently needed to solve this problem. Yang Priscilla L. et al. synthesized many NS3-targeting PROTACs [39] by linking VX-950 and CRBN Binder based on the PROTAC strategy. Among them, the representative compound PROTAC 70 (Table 2) can effectively degrade NS3 in human hepatoma adherent Huh7.5 cells. In addition, PROTAC 70 can also degrade V55A and A156S mutant NS3. Therefore, the successful discovery of this degradation agent is a boon for HCV-infected patients. This study also suggests that PROTACs may also be potential antiviral drugs, a strategy that has also been used to target SARS-CoV-2 as it emerges. The major proteases (Mpro and PLpro) [40] and RNA-dependent RNA polymerase (RdRP) [41] of SARS-CoV-2 are currently targeted by small molecule inhibitors [42]. They could be potential targets for PROTAC molecules.
Table 2. Representative PROTACs for antiviral.
| Indication | PROTAC | Target | Structure | Activity | Ref. | |
|---|---|---|---|---|---|---|
| DC50 | Dmax% | |||||
| HCV | 70 | NS3 | 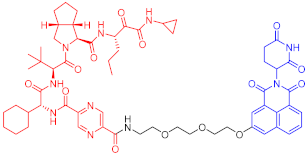 |
50 nM | - | [39] |
Note: red: molecule to bind to POI, black: linker, blue: ligand of E3 ligase.
References
- Alabi, S.B.; Crews, C.M. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647.
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559.
- Gu, S.; Cui, D.; Chen, X.; Xiong, X.; Zhao, Y. PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery. BioEssays 2018, 40, e1700247.
- Yang, J.; Li, Y.; Aguilar, A.; Liu, Z.; Yang, C.Y.; Wang, S. Simple Structural Modifications Converting a Bona fide MDM2 PROTAC Degrader into a Molecular Glue Molecule: A Cautionary Tale in the Design of PROTAC Degraders. J. Med. Chem. 2019, 62, 9471–9487.
- Tan, L.; Gray, N.S. When Kinases Meet PROTACs. Chin. J. Chem. 2018, 36, 971–977.
- Scheepstra, M.; Hekking, K.F.W.; van Hijfte, L.; Folmer, R.H.A. Bivalent Ligands for Protein Degradation in Drug Discovery. Comput. Struct. Biotechnol. J. 2019, 17, 160–176.
- Sakamoto, K.M. Protacs for treatment of cancer. Pediatr. Res. 2010, 67, 505–508.
- Ottis, P.; Crews, C.M. Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chem. Biol. 2017, 12, 892–898.
- Nguyen, C.; West, G.M.; Geoghegan, K.F. Emerging Methods in Chemoproteomics with Relevance to Drug Discovery. Methods Mol. Biol. 2017, 1513, 11–22.
- Raina, K.; Crews, C.M. Chemical inducers of targeted protein degradation. J. Biol. Chem. 2010, 285, 11057–11060.
- Nowak, R.P.; DeAngelo, S.L.; Buckley, D.; He, Z.; Donovan, K.A.; An, J.; Safaee, N.; Jedrychowski, M.P.; Ponthier, C.M.; Ishoey, M.; et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 2018, 14, 706–714.
- An, S.; Fu, L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 2018, 36, 553–562.
- Farnaby, W.; Koegl, M.; Roy, M.J.; Whitworth, C.; Diers, E.; Trainor, N.; Zollman, D.; Steurer, S.; Karolyi-Oezguer, J.; Riedmueller, C.; et al. Publisher Correction: BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019, 15, 846.
- Drummond, M.L.; Williams, C.I. In Silico Modeling of PROTAC-Mediated Ternary Complexes: Validation and Application. J. Chem. Inf. Modeling 2019, 59, 1634–1644.
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521.
- Roy, M.J.; Winkler, S.; Hughes, S.J.; Whitworth, C.; Galant, M.; Farnaby, W.; Rumpel, K.; Ciulli, A. SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem. Biol. 2019, 14, 361–368.
- Hughes, S.J.; Ciulli, A. Molecular recognition of ternary complexes: A new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017, 61, 505–516.
- Riching, K.M.; Mahan, S.; Corona, C.R.; McDougall, M.; Vasta, J.D.; Robers, M.B.; Urh, M.; Daniels, D.L. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Biol. 2018, 13, 2758–2770.
- Toure, M.; Crews, C.M. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew. Chem. Int. Ed. Engl. 2016, 55, 1966–1973.
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229.
- Chen, Z.J.; Sun, L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 2009, 33, 275–286.
- Jevtić, P.; Haakonsen, D.L.; Rapé, M. An E3 ligase guide to the galaxy of small-molecule-induced protein degradation. Cell Chem. Biol. 2021, 28, 1000–1013.
- Kannt, A.; Đikić, I. Expanding the arsenal of E3 ubiquitin ligases for proximity-induced protein degradation. Cell Chem. Biol. 2021, 28, 1014–1031.
- Bond, M.J.; Crews, C.M. Proteolysis targeting chimeras (PROTACs) come of age: Entering the third decade of targeted protein degradation. RSC Chem. Biol. 2021, 2, 725–742.
- Bemis, T.A.; La Clair, J.J.; Burkart, M.D. Unraveling the Role of Linker Design in Proteolysis Targeting Chimeras. J. Med. Chem. 2021, 64, 8042–8052.
- Benowitz, A.B.; Jones, K.L.; Harling, J.D. The therapeutic potential of PROTACs. Expert Opin. Ther. Pat. 2021, 31, 1–24.
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019, 4, 64.
- Zeng, S.; Huang, W.; Zheng, X.; Liyan, C.; Zhang, Z.; Wang, J.; Shen, Z. Proteolysis targeting chimera (PROTAC) in drug discovery paradigm: Recent progress and future challenges. Eur. J. Med. Chem. 2021, 210, 112981.
- Neklesa, T.; Snyder, L.B.; Willard, R.R.; Vitale, N.; Raina, K.; Pizzano, J.; Gordon, D.A.; Bookbinder, M.; Macaluso, J.; Dong, H.; et al. An oral androgen receptor PROTAC degrader for prostate cancer. J. Clin. Oncol. 2018, 36, 381.
- Flanagan, J.J.; Qian, Y.; Gough, S.M.; Andreoli, M.; Bookbinder, M.; Cadelina, G.; Bradley, J.; Rousseau, E.; Willard, R.; Pizzano, J.; et al. Abstract P5-04-18: ARV-471, an oral estrogen receptor PROTAC degrader for breast cancer. Cancer Res. 2019, 79, P5–P04.
- Kim, J.; Kim, H.; Park, S.B. Privileged structures: Efficient chemical “navigators” toward unexplored biologically relevant chemical spaces. J. Am. Chem. Soc. 2014, 136, 14629–14638.
- Xi, M.; Chen, Y.; Yang, H.; Xu, H.; Du, K.; Wu, C.; Xu, Y.; Deng, L.; Luo, X.; Yu, L.; et al. Small molecule PROTACs in targeted therapy: An emerging strategy to induce protein degradation. Eur. J. Med. Chem. 2019, 174, 159–180.
- Baigent, C.; Keech, A.; Kearney, P.M.; Blackwell, L.; Buck, G.; Pollicino, C.; Kirby, A.; Sourjina, T.; Peto, R.; Collins, R.; et al. Efficacy and safety of cholesterol-lowering treatment: Prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 2005, 366, 1267–1278.
- Stone, N.J.; Robinson, J.G.; Lichtenstein, A.H.; Bairey Merz, C.N.; Blum, C.B.; Eckel, R.H.; Goldberg, A.C.; Gordon, D.; Levy, D.; Lloyd-Jones, D.M.; et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014, 63, 2889–2934.
- Li, M.X.; Yang, Y.; Zhao, Q.; Wu, Y.; Song, L.; Yang, H.; He, M.; Gao, H.; Song, B.L.; Luo, J.; et al. Degradation versus Inhibition: Development of Proteolysis-Targeting Chimeras for Overcoming Statin-Induced Compensatory Upregulation of 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase. J. Med. Chem. 2020, 63, 4908–4928.
- Luo, G.; Li, Z.; Lin, X.; Li, X.; Chen, Y.; Xi, K.; Xiao, M.; Wei, H.; Zhu, L.; Xiang, H. Discovery of an orally active VHL-recruiting PROTAC that achieves robust HMGCR degradation and potent hypolipidemic activity in vivo. Acta Pharm. Sin. B 2021, 11, 1300–1314.
- Choo, Q.L.; Kuo, G.; Weiner, A.J.; Overby, L.R.; Bradley, D.W.; Houghton, M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989, 244, 359–362.
- Schulze zur Wiesch, J.; Schmitz, H.; Borowski, E.; Borowski, P. The proteins of the Hepatitis C virus: Their features and interactions with intracellular protein phosphorylation. Arch. Virol. 2003, 148, 1247–1267.
- De Wispelaere, M.; Du, G.; Donovan, K.A.; Zhang, T.; Eleuteri, N.A.; Yuan, J.C.; Kalabathula, J.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019, 10, 3468.
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and COVID-19 Therapeutics. ChemMedChem 2020, 15, 907–932.
- Yin, W.; Mao, C.; Luan, X.; Shen, D.D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504.
- Boras, B.; Jones, R.M.; Anson, B.J.; Arenson, D.; Aschenbrenner, L.; Bakowski, M.A.; Beutler, N.; Binder, J.; Chen, E.; Eng, H.; et al. Discovery of a Novel Inhibitor of Coronavirus 3CL Protease for the Potential Treatment of COVID-19. bioRxiv 2021.
More
Information
Subjects:
Chemistry, Medicinal
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
3 times
(View History)
Update Date:
26 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





