Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Luisa Di Stefano | -- | 1612 | 2022-09-22 11:03:00 | | | |
| 2 | Jessie Wu | + 1 word(s) | 1613 | 2022-09-23 07:10:16 | | | | |
| 3 | Jessie Wu | Meta information modification | 1613 | 2022-09-23 07:12:12 | | | | |
| 4 | Jessie Wu | Meta information modification | 1613 | 2022-09-23 07:13:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Stefano, L.D. Classes of Transposable Elements. Encyclopedia. Available online: https://encyclopedia.pub/entry/27468 (accessed on 28 February 2026).
Stefano LD. Classes of Transposable Elements. Encyclopedia. Available at: https://encyclopedia.pub/entry/27468. Accessed February 28, 2026.
Stefano, Luisa Di. "Classes of Transposable Elements" Encyclopedia, https://encyclopedia.pub/entry/27468 (accessed February 28, 2026).
Stefano, L.D. (2022, September 22). Classes of Transposable Elements. In Encyclopedia. https://encyclopedia.pub/entry/27468
Stefano, Luisa Di. "Classes of Transposable Elements." Encyclopedia. Web. 22 September, 2022.
Copy Citation
Transposable elements (TEs) are mobile genetic elements that constitute a sizeable portion of many eukaryotic genomes. Through their mobility, they represent a major source of genetic variation, and their activation can cause genetic instability and has been linked to aging, cancer and neurodegenerative diseases.
chromatin
transposable elements
DNA
1. Introduction
Eukaryotic genomes are historical records of transposable element (TE) integration and mobilization events that occurred over millions of years. TEs and their remnants (degenerated TE sequences) represent a large fraction of eukaryotic genomes, constituting approximately half of the human genome. Transposable elements are mostly repetitive DNA sequences, and as their name indicates, they are capable of moving within the genome. Once considered “junk” DNA, it is now clear that TEs can both negatively and positively impact their host genomes (Figure 1A). TEs can threaten genomic stability through their ability to move around the genome; the insertion of a TE into a coding gene or a gene regulatory element impacts gene structure and expression and can lead to diseases such as cancer, hemophilia or neurodegenerative disorders (Figure 1B) [1][2]. TEs can also trigger chromosome deletions, duplications, inversions and translocations through ectopic recombination between TEs belonging to the same family [3] (Figure 1C). TE-driven genomic rearrangements have been responsible for major genomic expansions, and there is evidence that they have contributed to speciation [4][5][6]. Intact TEs can code for proteins that allow them to hop within the genome; however, most TE sequences degenerate over time and lose this ability [7]. Nevertheless, they can still play important roles in the host genome. Some have become host cell genes, a phenomenon known as TE domestication. This is the case of the Syncytin genes involved in placental development [8] (Figure 1A,D). In addition to producing coding transcripts, some TEs can be transcribed to produce non-coding RNAs. These non-coding RNAs can exert specific biological functions, as is the case of a transcript produced by the LINE-1 retrotransposon that works as an RNA scaffold during mouse early developmental stages [9] (Figure 1E). Additionally, a growing body of evidence shows that TE sequences have been co-opted to serve as regulatory elements to host genes. TEs carry cis-regulatory elements that, by duplication and insertion, can redistribute transcription factor binding sites and alter gene expression patterns. Epigenomic analyses indicate that a large fraction of mammalian regulatory binding sites (promoters/enhancers) have been provided by TE-derived sequences (Figure 1F) [10][11]. TE derived cis-regulatory elements can also influence chromatin architecture by serving as binding sites for CTCF (the CCCTC-binding factor), a sequence-specific DNA-binding protein that contributes to the establishment of chromatin loops [12][13]. Sometimes, TE presence itself can regulate host genome expression by modifying chromatin accessibility. For example, TEs can act as heterochromatin nucleation centers by inducing the spread of silencing marks from the TE to the adjacent cis-regulatory elements of host genes, thus inducing their repression (Figure 1G) [14]. However, TEs can also create de novo insulator regions, shielding a gene from heterochromatin expansion and allowing its expression [15][16][17].
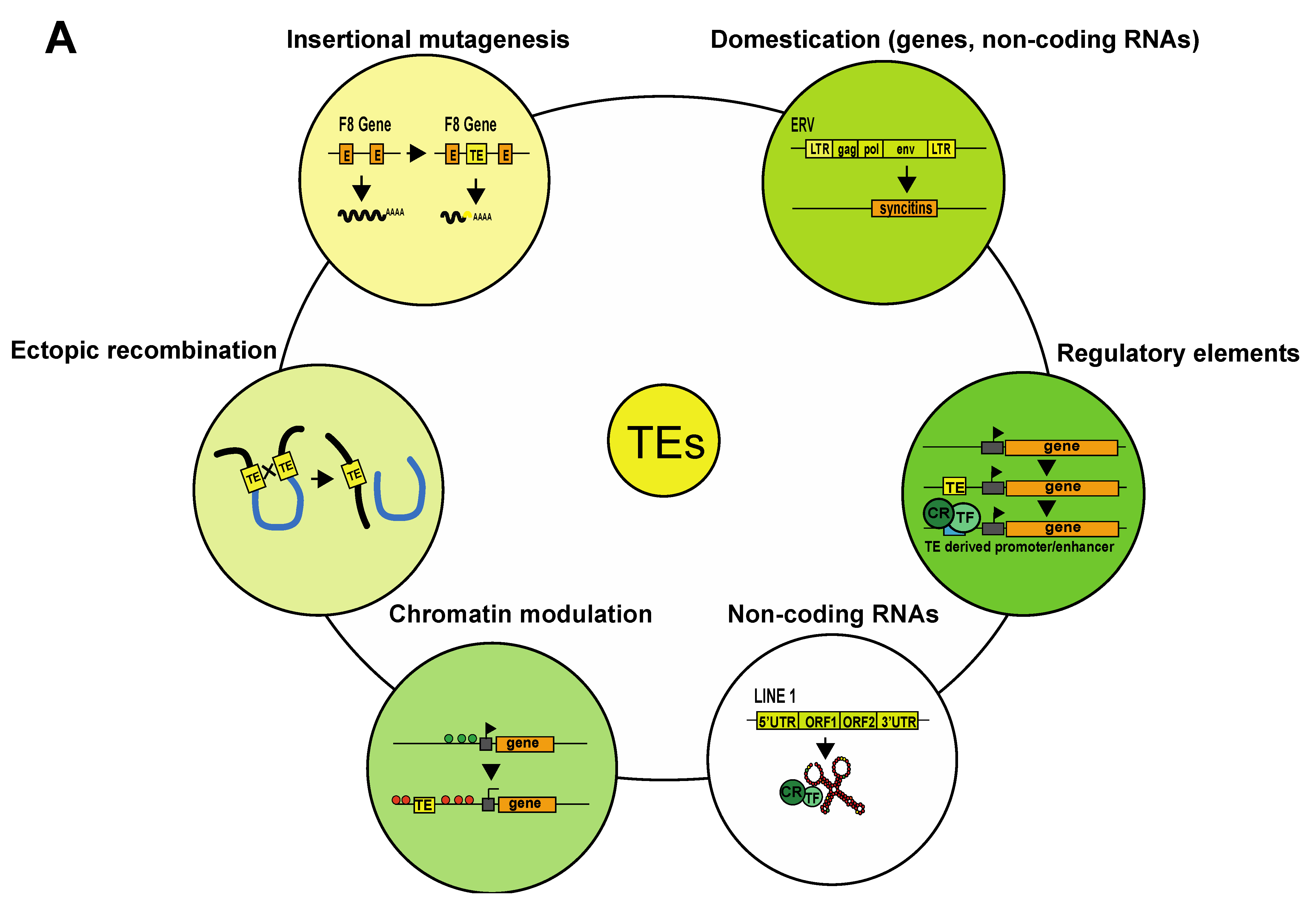
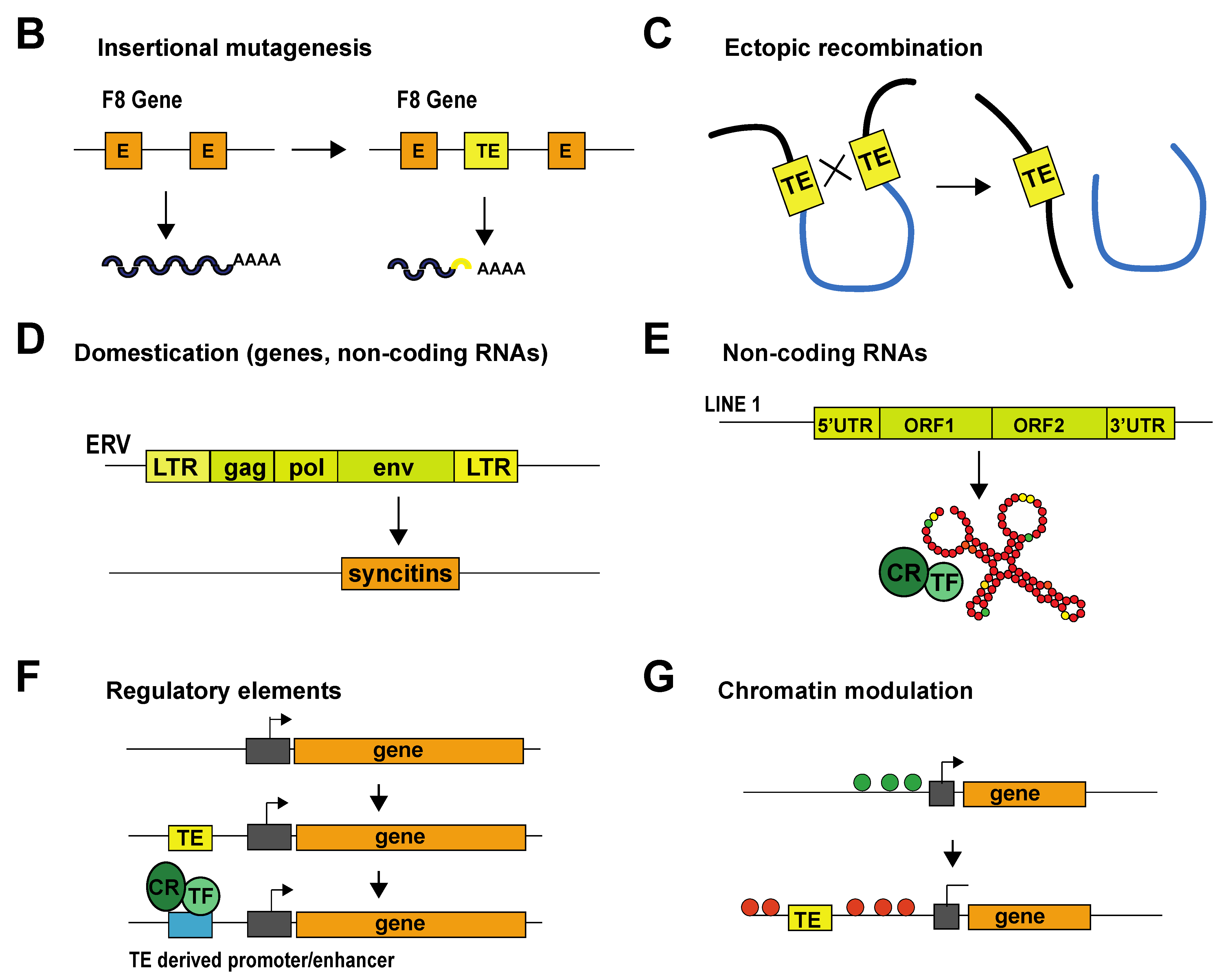
Figure 1. Impact of TEs on their host genome. (A) Examples of how TEs can impact genomes. (B) Schematic representation of a how insertion of a transposable element (TE) into the open reading frame of the coagulation factor VIII (F8) gene can induce insertional mutagenesis. This mutation was found in patients with hemophilia [1]. (C) Schematic representation of TE-induced ectopic recombination. (D) An example of TE domestication. Ancient env genes from ERVs have evolved into syncytin genes, which are involved in placenta formation [8]. Another example not represented here is that of Rag1 and Rag2, which are involved in V(D)J somatic recombination in the immune system of vertebrates [18]. (E) An example of a TE transcript (LINE1) acting as an RNA scaffold for chromatin regulators and transcription factors. (F) TE sequences carry transcription factor binding sites, and their insertion can lead to novel gene-regulatory patterns in the host organism. (G) Example of how a TE can modulate chromatin by inducing the spread of heterochromatin. Abbreviations: E, exon; TE, transposable element; LTR, long terminal repeat; ORF, open reading frame; UTR, untranslated region; CR, chromatin regulator; TF, transcription factor.
In summary, TEs, through their capacity to impact gene expression patterns and induce genome instability, are an important source of genetic variation and a driving force of genomic evolution.
2. Classes of Transposable Elements
Transposable elements are broadly classified on the basis of their mechanism of transposition as class I elements (retrotransposons) and class II elements (DNA transposons). Class I elements are transcribed into an RNA intermediate and use reverse transcriptase to form a new copy of their DNA, which is then inserted into the host genome (copy and paste) (Figure 2A). Class I elements are subdivided, on the basis of the presence or absence of long terminal repeats (LTRs), into LTR and non-LTR elements (Box 1). For LTR elements, the retrotranscription occurs in cytoplasmic virus-like particles, and the resulting dsDNA is then imported into the nucleus, where an integrase inserts it into the host genome. For non-LTR retrotransposons, retrotranscription occurs at the target locus of the host genome, a process known as ‘target-primed reverse transcription’ [19]. Class II elements encode a transposase enzyme that excises the parental sequence from a donor site and reintegrates it into another location in the genome (cut and paste) (Figure 2B). Therefore, in contrast to retrotransposons, they generally do not accumulate in copy number. However, they have adopted strategies to increase in copy number by transposing during host DNA synthesis from replicated to unreplicated sites or by taking advantage of the error-prone homologous recombination repair process [20]. More recently, rolling-circle elements (e.g., Helitrons) have been identified as a distinct group of abundant DNA transposons that do not replicate via the “cut-and-paste” mechanism but through a “peel-and-paste” mechanism. It has been hypothesized that the sense strand is “peeled” off, serving as a template to synthesize a second strand to form a circular double-stranded DNA (dsDNA) intermediate (Figure 2B) [20].
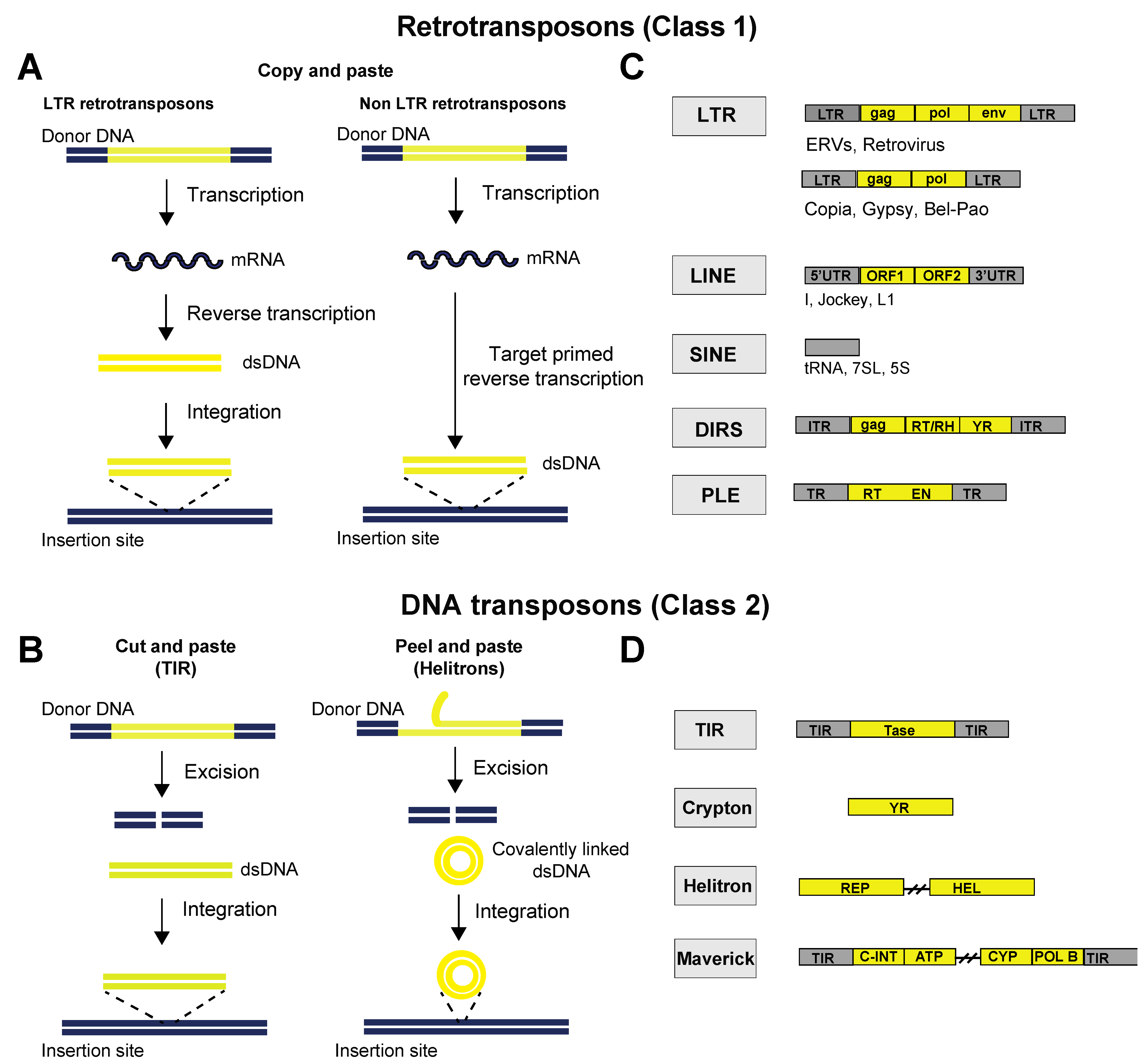
Figure 2. Schematic representation of the mobilization mechanisms of transposable elements. (A) Schematic representation of the “copy-and-paste” mobilization mechanism of retrotransposons. Retrotransposons replicate through an RNA intermediate and a reverse transcription step. LTR retrotransposons produce a double-stranded DNA (dsDNA) intermediate that integrates into a new locus, whereas non-LTR retrotransposons retrotranscribe directly at the target locus after cleaving genomic DNA, a process known as ‘target-primed reverse transcription’. (B) Schematic representation of the “cut-and-paste” and “peel-and-paste” mobilization mechanisms of DNA transposons. Both mobilization mechanisms require the excision of the transposon DNA from its original locus and its reintegration into another locus, but the “peel-and-paste” mechanism requires the formation of a circular double-stranded DNA (dsDNA) intermediate. The mechanism of replication of maverick and crypton elements has not been determined. (C,D) Classification of eukaryotic transposable elements (as proposed by Wicker et al. [21][22]). Genetic structures of representative transposable elements from each order. Yellow boxes represent open reading frames (ORFs), and grey boxes represent non-coding domains. Element lengths are not to scale. Abbreviations: LTR, long terminal repeat; ORF, open reading frame; UTR, untranslated region; ENV, envelope protein; GAG, capsid protein; RT, reverse transcriptase; RH, ribonuclease H domain; ITR, inverted terminal repeat; TR, terminal repeat; EN, endonuclease; YR, tyrosine recombinase; TIR, terminal inverted repeats; Tase, transposase; REP, replication initiator; Hel, helicase; C-INT, integrase; ATP, packaging ATPase; CYP, cysteine protease; POL B, DNA polymerase B.
Independent from their mechanism of amplification, transcription is a crucial step in the replication of all groups of transposons. In the case of retrotransposons, RNA serves as a template for both the translation of TE proteins and for reverse transcription. For DNA transposons, transcription allows the expression of the transposase, which is essential for mobilization. These observations underline the need to fully understand the mechanisms underlying TE transcriptional regulation.
Box 1. Classification of Transposable Elements.
|
The classification of TEs is constantly being updated thanks to the development of novel tools that allow for a more refined TE classification and the discovery of new TE types. Traditionally, TEs have been classified into two classes on the basis of the DNA or RNA intermediate of their element: retrotransposons (class 1) and DNA transposons (class 2) (Figure 2A,B). Retrotransposons can be further classified into five orders based on their structural organization and mechanistic aspects of replication: long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), DIRS-like elements (DIRSs) and Penelope-like elements (PLEs) (Figure 2C). LTR elements are characterized by the presence of 5’ and 3’ non-coding long terminal repeat sequences that control the expression of retroviral genes. LINEs contain a 5’UTR and a polyA signal and encode all the proteins necessary for retrotransposition. SINEs are non-autonomous elements, the retrotransposition of which relies on functions coded by coexisting LINEs. DIRS-like elements have diverged from the other retrotransposons because they do not possess an integrase (INT) but rather use a tyrosine recombinase (YR) to integrate in the host genome. PLEs harbor an ORF coding for a protein that contains reverse transcriptase (RT) and endonuclease (EN) domains. PLEs are absent from mammalian genomes but can be found in some other eukaryotic genomes, including Drosophila, where they can cause hybrid dysgenesis syndrome, which is characterized by simultaneous mobilization of several unrelated TE families in the progeny of crosses involving different strains of the same species.
DNA transposons (class 2) are subdivided into the following orders: terminal inverted repeats (TIRs), Cryptons, Helitrons and Mavericks (Figure 2D). TIRs are characterized by the presence of terminal inverted repeats (TIRs) and encode a transposase that mediates excision and integration through binding to TIRs. Cryptons are simple transposons consisting in a single ORF coding for a tyrosine recombinase (YR). Helitrons code for a helicase. They replicate via the “peel-and-paste” mechanism by forming a circular double-stranded DNA (dsDNA) intermediate, earning the name of rolling-circle transposons. Mavericks are large DNA transposons encoding various proteins, including a DNA polymerase and an integrase.
|
References
- Kazazian, H.H., Jr.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166.
- Hancks, D.C.; Kazazian, H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9.
- Robberecht, C.; Voet, T.; Zamani Esteki, M.; Nowakowska, B.A.; Vermeesch, J.R. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013, 23, 411–418.
- Naville, M.; Henriet, S.; Warren, I.; Sumic, S.; Reeve, M.; Volff, J.N.; Chourrout, D. Massive Changes of Genome Size Driven by Expansions of Non-autonomous Transposable Elements. Curr. Biol. 2019, 29, 1161–1168.e6.
- Shao, F.; Han, M.; Peng, Z. Evolution and diversity of transposable elements in fish genomes. Sci. Rep. 2019, 9, 15399.
- Serrato-Capuchina, A.; Matute, D.R. The Role of Transposable Elements in Speciation. Genes 2018, 9, 254.
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199.
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789.
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e19.
- Goerner-Potvin, P.; Bourque, G. Computational tools to unmask transposable elements. Nat. Rev. Genet. 2018, 19, 688–704.
- Morgan, H.D.; Sutherland, H.G.; Martin, D.I.; Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999, 23, 314–318.
- Gualdrini, F.; Polletti, S.; Simonatto, M.; Prosperini, E.; Pileri, F.; Natoli, G. H3K9 trimethylation in active chromatin restricts the usage of functional CTCF sites in SINE B2 repeats. Genes Dev. 2022, 36, 414–432.
- Kaaij, L.J.T.; Mohn, F.; van der Weide, R.H.; de Wit, E.; Buhler, M. The ChAHP Complex Counteracts Chromatin Looping at CTCF Sites that Emerged from SINE Expansions in Mouse. Cell 2019, 178, 1437–1451.e14.
- Sentmanat, M.F.; Elgin, S.C. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc. Natl. Acad. Sci. USA 2012, 109, 14104–14109.
- Etchegaray, E.; Naville, M.; Volff, J.N.; Haftek-Terreau, Z. Transposable element-derived sequences in vertebrate development. Mob. DNA 2021, 12, 1.
- Ninova, M.; Chen, Y.A.; Godneeva, B.; Rogers, A.K.; Luo, Y.; Fejes Toth, K.; Aravin, A.A. Su(var)2-10 and the SUMO Pathway Link piRNA-Guided Target Recognition to Chromatin Silencing. Mol. Cell 2020, 77, 556–570.e556.
- Lee, Y.C.G.; Karpen, G.H. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 2017, 6, e25762.
- Kapitonov, V.V.; Koonin, E.V. Evolution of the RAG1-RAG2 locus: Both proteins came from the same transposon. Biol. Direct 2015, 10, 20.
- Lanciano, S.; Cristofari, G. Measuring and interpreting transposable element expression. Nat. Rev. Genet. 2020, 21, 721–736.
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561.
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982.
- Makalowski, W.; Gotea, V.; Pande, A.; Makalowska, I. Transposable Elements: Classification, Identification, and Their Use As a Tool For Comparative Genomics. Methods Mol. Biol. 2019, 1910, 177–207.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
5.3K
Revisions:
4 times
(View History)
Update Date:
23 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





