Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Angelantonio Maglio | -- | 3597 | 2022-09-07 19:30:58 | | | |
| 2 | Catherine Yang | Meta information modification | 3597 | 2022-09-08 03:25:38 | | | | |
| 3 | Catherine Yang | Meta information modification | 3597 | 2022-09-09 05:52:43 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Vatrella, A.; Maglio, A.; Pelaia, C.; Ciampo, L.; Pelaia, G.; Vitale, C. Eosinophil Structure and Biology. Encyclopedia. Available online: https://encyclopedia.pub/entry/26976 (accessed on 02 March 2026).
Vatrella A, Maglio A, Pelaia C, Ciampo L, Pelaia G, Vitale C. Eosinophil Structure and Biology. Encyclopedia. Available at: https://encyclopedia.pub/entry/26976. Accessed March 02, 2026.
Vatrella, Alessandro, Angelantonio Maglio, Corrado Pelaia, Luigi Ciampo, Giulia Pelaia, Carolina Vitale. "Eosinophil Structure and Biology" Encyclopedia, https://encyclopedia.pub/entry/26976 (accessed March 02, 2026).
Vatrella, A., Maglio, A., Pelaia, C., Ciampo, L., Pelaia, G., & Vitale, C. (2022, September 07). Eosinophil Structure and Biology. In Encyclopedia. https://encyclopedia.pub/entry/26976
Vatrella, Alessandro, et al. "Eosinophil Structure and Biology." Encyclopedia. Web. 07 September, 2022.
Copy Citation
Eosinophils are granulocytes with unique biology. The fact that these cells have been largely preserved during evolution strongly suggests that they play relevant physiological functions. Eosinophils have traditionally been classified as effector cells with prevalent cytotoxic activity, although recent evidence indicates that these cells may play a role in a wide range of homeostatic and regulatory functions.
eosinophil
severe asthma
type 2 inflammation
molecular biology
cell biology
allergy
1. Eosinophil Structure
Eosinophils are polymorphonuclear leukocytes, usually measuring 10–16 μm in diameter, with a segmented bilobed nucleus. Characteristic of this cell is the presence of a large number of molecules with pleiotropic functions, such as cationic granule proteins, chemokines, cytokines, growth factors, immunomodulatory molecules, lipid mediators, mainly accumulated within the intracellular compartment (Figure 1). Eosinophils also have a large array of transmembrane proteins (integrins) and surface receptors which mediate the interaction with the micro-environment and allow the response to multiple stimuli (Figure 1) [1].
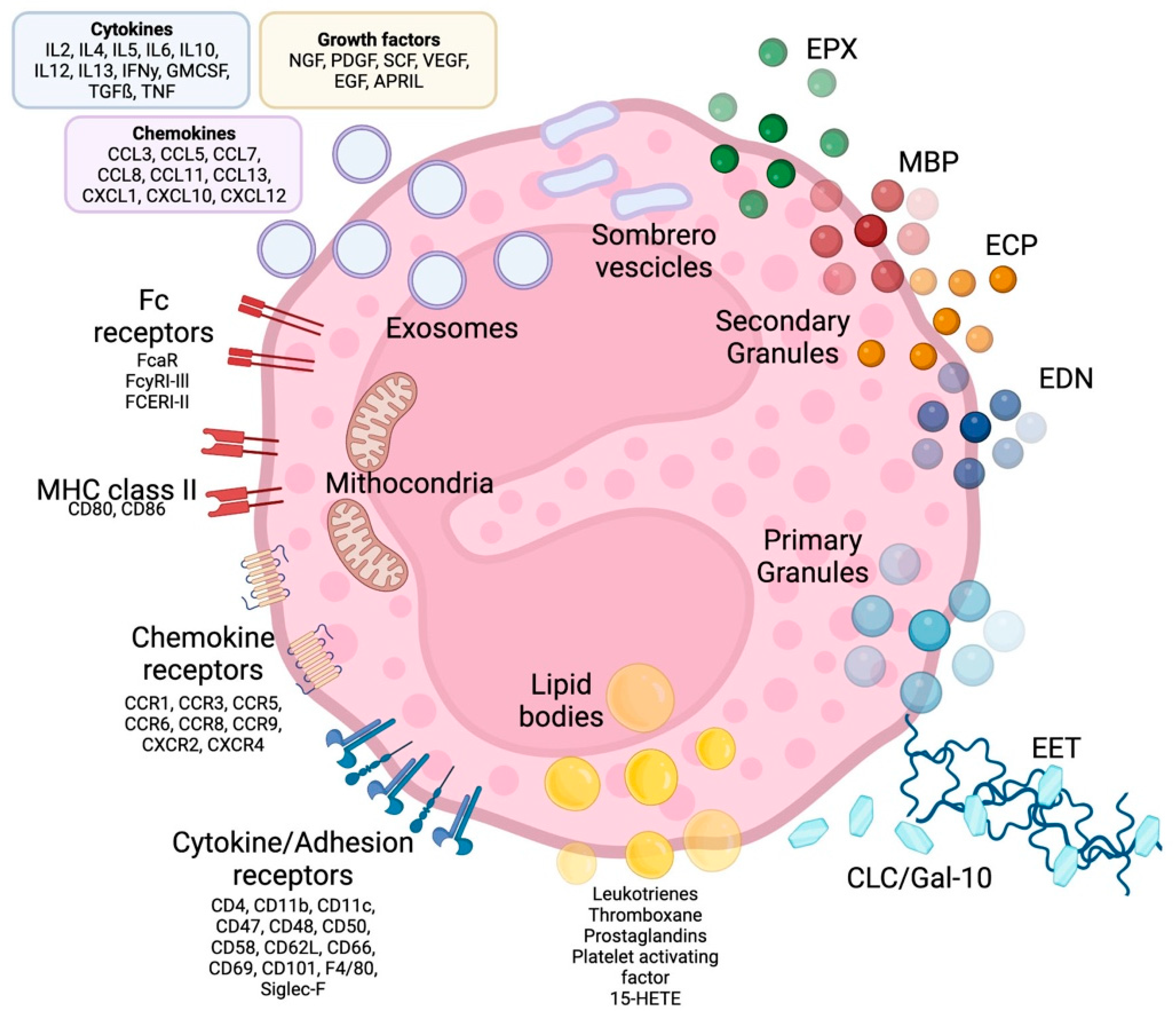
Figure 1. Eosinophil structure, granules, surface receptors, exosomes and EETs. Eosinophils are supplied by a large number of proteins, receptors and enzymes that allow them to interact with the microenvironment and express a number of receptors on their surface, including receptors for cytokines, chemokines and lipid mediators, which are involved in cell growth, survival, adhesion, migration and activation. In addition to receptors, adhesion molecules such as integrins are expressed on the cell surface, which allows eosinophils to migrate and react to several stimuli. The effects of eosinophils are largely achieved due to the content of their granules. Primary granules include Charcot-Leyden/galectin-10 protein, a characteristic eosinophilic protein implicated in asthma and parasitic infections, as well as a constituent part of so-called eosinophilic extracellular traps, whose other major constituents are nuclear or mitochondrial DNA strands. Specific or secondary granules contain four main cationic proteins: MBP, ECP, EPX and EDN. In addition, some of the content of the granules is released through particular vesicles called sombrero vesicles. Each of them has different effects, clarified in the text. Lipid bodies contain prostaglandins, thromboxane and leukotrienes, which participate in allergic inflammation, fibrosis and thrombosis. Finally, eosinophils are able to release exosomes that fuse with the cell membrane, which are involved in epithelial damage. CLC/Gal-10: Charcot-Leyden crystal proteins; ECP: eosinophil cationic protein; MBP: major basic proteins; EPX: eosinophil peroxidase; EDN: eosinophil-derived neurotoxin; MHC class II: Mayor histocompatibility complex-II; EET: eosinophilic extracellular traps. See the text for further explanation.
1.1. Eosinophil Granules
Eosinophil cytoplasm is packed with different types of granules (Figure 1). The two major types of large granules present within mature human eosinophils are specific granules and immature specific granules. The specific granules, also called “secondary granules”, consist of a dense crystalline nucleus surrounded by a membrane, a unique morphology found only in eosinophils. [2]; these granules contain a large variety of mediators, including basic proteins, cytokines, chemokines, growth factors and enzymes, which are able to produce tissue inflammation and damage. The main represented specific granule substances are eosinophil cationic protein (ECP), major basic proteins (MBP-1 and MBP-2), eosinophil peroxidase (EPX) and eosinophil-derived neurotoxin (EDN) [3]. Specific immature granules, also called “primary granules” are tendentially smaller than specific granules and are the principal location of Charcot-Leyden crystal (CLC) protein (a member of the carbohydrate-binding family of galectin-10).
In eosinophils have also been identified a third intracellular compartment, the lipid bodies, is specifically committed to the production of eicosanoid mediators of inflammation.
Eosinophil sombrero vesicles (EoSVs) are not granules, but distinct tubular vesicles that tend to curl into a hoop-like morphology, giving rise to the term. EoSVs derive from specific granules and travel to the cell membrane to discharge their contents to the extracellular domain.
1.2. Eosinophil Surface Receptors
Eosinophils display a vast array of receptors and surface molecules, which allow them to integrate with the innate and adaptive branches of the immune system involved in inflammatory responses and homeostasis. While many are selectively expressed on eosinophils such as interleukin-5Rα, CC-chemokine receptor 3 (CCR3), sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8), the epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1) appears completely unique to the eosinophil. The wide range of receptors present on eosinophils makes theme very versatile cells, with the ability to react to the stimulus, co-stimulate cells in antigen presentation and migrate to tissues in both physiological and pathological conditions [1].
-
Cytokine Receptors
Eosinophils display receptors for IL-3, IL-5, and granulocyte–macrophage colony-stimulating factor (GM-CSF), the three main cytokines involved in differentiation and maturation of these cells. The heterodimeric receptor for IL-5 is likely to be the most important cytokine receptor expressed by eosinophils, since IL-5 plays a fundamental role in all stages of eosinophil biology. The alpha-subunit, IL-5Rα, is specific to IL-5, while the beta-subunit is shared with the receptors for IL-3 and GM-CSF. Eosinophils also possess specific receptors for various other cytokines and growth factors, including IL-4, IL-13, IL-33, thymic stromal lymphopoietin (TSLP), and transforming growth factor-β (TGF-β) [2].
-
Adhesion Receptors
Eosinophils express various types of membrane adhesion receptors, primarily integrins and selectins, which are up regulated by a wide range of pro-inflammatory cytokines and chemokines.
Integrin molecules are trans-membrane glycoproteins made up of an α and a β chain that includes the very late antigen-4 (VLA-4, CD49d/CD29) and the complement receptor CR3 (CD11b/CD18), also known as macrophage-1 (Mac-1) antigen.
Selectins are surface glycoproteins belonging to three groups (E-, L-, and P-selectin), in particular, L-selectin (CD62L) and P-selectin glycoprotein ligand-1 (CD162) are constitutively and highly expressed on circulating eosinophils [4].
Adhesion molecules act in a coordinated way, allowing eosinophils to roll and adhere to endothelia, thus facilitating their migration and accumulation at the sites of inflammation.
-
Chemoattractant Receptors
Eosinophils express on their surface various seven-transmembrane spanning G protein-coupled receptors for chemokines. Among them, CCR3 is an important, highly expressed receptor that binds to all three subtypes of eotaxin (a selective eosinophilic chemo-attractant) and to other chemokines, including the monocyte-3 chemoattractive protein (MCP-3) and MCP-4. The relevant role of CCR3 in asthma pathology is also supported by the evidence that the airways of patients with asthma contain more cells expressing mRNA for CCR3 and its ligands than non-asthmatic controls [5]. CCR1 is another key chemokine receptor on the surface of eosinophils, activated by chemoattractant cytokine ligand-3 (CCL-3) and CCL-5 (also known as RANTES: regulated on activation, normal T cell expressed and secreted).
-
Fc Receptors
The eosinophil displays various immunoglobulin (Ig) receptors and related family members involved in functional activities in which eosinophils are involved, including antibody-mediated cellular cytotoxicity (ADCC) for helminths and other immunomodulatory functions and pathological activities in diseases associated with eosinophilia. Fc receptors for IgA, IgD, IgE, IgG and IgM, localized on the membrane of eosinophils, promote interaction with the adaptive immune system.
The high-affinity Fc-epsilon R1-alpha (FcεR1) binding IgE is usually expressed in very small quantities in a trimeric form (without a β chain) and seems to have no role in eosinophil activation [6]. In contrast, cross-linking of FcαRI and FcγRII, with IgA and IgG, respectively, has been shown to induce eosinophil activation [7].
-
Major Histocompatibility Complex-II
Eosinophils express major histocompatibility complex class II (MHC-II) and co-stimulatory molecules such as CD80 and CD86, necessary for T-cell activation and proliferation. Lung eosinophils of asthmatic patients undergoing allergen challenges express higher levels of HLA-DR (a subtype of the MHC-II molecule) than blood eosinophils [8].
-
Pattern Recognition Receptors (PRRs)
Pattern Recognition Receptors are membrane proteins expressed on the surface of eosinophils that are directly stimulated during host innate immune responses from pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs); these PRRs promote the interactions of eosinophils with invading microorganisms (especially helminths) and with the surrounding microenvironment. Among a variety of homeostatic and anti-infective activities, PRRs regulate the immune response and tissue damage [9]. Toll-like receptors (TLRs) are one of the most represented subtypes of PRR expressed by eosinophils (as well as many other cell lines) on their surface and even on endosomes [10]. Other significant PRRs expressed by eosinophils also include proteinase-activated receptors PAR-1 and -2. The latter could play a relevant role in the activation of eosinophils in response to proteases released by aeroallergens such as dust mites, fungi, or pollen [2]; moreover, there are other types of PRRs with partially overlapping characteristics of the previous mentioned, like retinoic acid-inducible gene-I-like receptors, nucleotide-binding oligomerisation domain-like (NOD-like) receptors, and the receptor for advanced glycation end products (RAGE) [8].
-
Lipid Mediator Receptors
Eosinophils express specific receptors for lipid mediators such leukotrienes, prostaglandins, and platelet-activating factors involved in eosinophil chemotaxis and transmigration.
-
Siglec-8
Sialic acid-binding immunoglobulin-like lectin (Siglec)-8 is an inhibitory receptor selectively expressed on human eosinophils, but information about its function in asthma pathology is still limited. Siglec-8 gene expression in asthma sputum cells is associated with type 2-high profiles of asthma and recent observation that administration of an antibody targeting Siglec-8 can induce selective eosinophil apoptosis, suggesting that it could represent a potential therapeutic target for eosinophil-mediated disease [11].
-
Inhibitory Receptors
Other inhibitory receptors regulating the survival and the activation of eosinophils, include CD300a, killer activating receptors, potassium inwardly-rectifying channel, and FcgRIIb [7].
1.3. Intracellular Receptors
The eosinophil has many intracellular receptors that regulate its function (such as some toll-like receptors and the glucocorticoid receptor). In the intracellular compartment of eosinophils, a splicing variant of the glucocorticoid receptor (GR-A) is present in large quantities [12]. GR-A is the pro-apoptotic isoform and is five times higher in eosinophils than in neutrophils, which is why eosinophils are much more susceptible than other cells to the therapeutic actions of glucocorticoids, such as apoptosis [13].
2. Eosinophil Biology
A series of sequential processes regulate the particular biology of eosinophils; these events occur in different compartments, from the bone marrow to the blood and peripheral tissues, in physiological or pathological conditions. All the different phases, from maturation to degranulation, are regulated by the interaction of the eosinophil with a series of molecules that include transcription factors, adhesion molecules and cytokines.
2.1. Eosinophil Differentiation and Maturation
Eosinophils are generated and developed in the bone marrow from multipotent hematopoietic stem cells, which create a population of committed progenitors of the eosinophilic lineage (EoPs) that in turn are capable of further differentiating into mature eosinophils, their terminal form [14].
Human EoPs are characterized by the expression of surface receptors such as CD34, CD38, and mainly high-affinity α subunit of the IL-5 receptor (IL-5Rα, or CD125) [15]. Differentiation of eosinophils normally occurs in the bone marrow; however, eosinophils can also develop from CD34+ EoPs which are found outside the bone marrow, blood and particularly lung tissue [16][17]. Increased levels of EoPs been identified in peripheral blood of atopic subjects compared to non-atopic controls [18]. Likewise, an increase in the number of CD34+/IL-5Rα+ EoPs has been identified in the bronchial mucosa of asthmatics compared to non-asthmatic controls [14]; moreover, the demonstration that blood EoPs have a greater response in vitro to IL-5 in patients with severe eosinophilic asthma than in milder asthmatics suggests a possible clinically relevant role of in situ eosinophilopoiesis in severe eosinophilic asthma [19].
Under homeostatic conditions, in healthy subjects, eosinophilopoiesis is mainly regulated by multiple transcription factors including GATA-binding protein 1 (GATA-1), Purine Rich Box-1 (PU.1), and the CCAAT-enhancing binding protein (c/EBP) family [20]. GATA-1 is thought to have the most important role, as disruption of the GATA-1 gene in mice results in a strain completely devoid of eosinophils [21].
The development of mature eosinophils in blood and peripheral tissues also depends on the synergistic contribution of cytokines such as IL-5, IL-3 and GM-CSF [2][22][23].
Eosinophils are fully differentiated after 7 days of maturation in the bone marrow. Mature cells are subsequently released in peripheral circulation, with a lifespan up to 24 h [24][25].
2.2. Eosinophil Migration and Activation
Under physiological conditions, the main migration site of eosinophils is the gastrointestinal tract from the stomach to the intestine. A minority of them also migrate to the lymph nodes, thymus, liver, spleen, uterus and mammary gland [25][26].
This recruitment in different tissues can take place as early as 8–12 h after entry into circulation, mainly through the binding of the CCR3 to different chemokines, such as CCL11 (eotaxin-1), CCL24 (eotaxin-2) and CCL26 (eotaxin-3) [26].
When within tissues, eosinophils can survive for up to 15 days, primarily exerting an immunomodulatory function, but also promoting tissue repair as well as antimicrobial and antifungal immunity [25][26]. In inflammatory conditions, eosinophils can infiltrate tissues where they are not normally found or are minimally present, such as large and small airways and esophagus [27], in consequence of a series of extremely well-established steps. In particular, during allergic inflammation and bronchial asthma, circulating eosinophils adhere to the vascular endothelium and roll along it before pouring into lung tissue. Initial contact with the endothelium depends on the binding of the eosinophil cell membrane P-selectin glycoprotein ligand-1 (PSGL-1) to the adhesion receptor P-selectin to the activated endothelium [11]. The binding of integrin very late antigen-4 (VLA-4) to the vascular cell adhesion molecule-1 (VCAM-1) promotes the activation and extravasation of eosinophils [11]. IL-13 causes increased eosinophilic expression of P-selectin and increased P-selectin-mediated adhesion to endothelial cells [2][28].
In patients with severe asthma EoPs can also migrate to airways, where they differentiate to mature cells in situ [19].
Under physiological conditions, activation, survival, and recruitment of eosinophils are largely driven by IL-5, a cytokine produced by type 2 helper T cells (Th2) that plays a prominent role in the regulation of eosinophils, EoPs, mast cells and type 2 innate lymphoid cells (ILC2). Indeed, IL-5 is a key regulator cytokine for eosinophils acting at multiple functional levels and time points during their lifespan. Epithelium-derived alarmins, including IL-33, IL-25, and thymic stromal lymphopoietin (TSLP) partly trigger the production of IL-5 [29][30][31][32].
In the context of eosinophilic asthma, the increase in eosinophils in the airways begins after exposure of the epithelium of the airways to various allergens or antigens, thus triggering the activation of an immunological cascade that directs eosinophils into the airways through the stimulation by Th2 cytokines and chemoattractants [33]. When helper T cells are activated by allergens, they switch to the Th2 phenotype and begin secreting IL-4, IL-5, and IL-13 [6][24].
IL-5 and RANTES are the most relevant inducers of eosinophil migration in the asthmatic lung [34]. The airway epithelium is also involved in the secretion and production of these Th2 cytokines through the production of IL-33, and IL-25, which are secreted after any type of epithelial insult [35][36][37]; these alarmins also activate ILC2s from the innate immune system, which also secrete and produce IL-5, IL-4, and IL-13 [38]; it is interesting to consider that the action of the epithelial alarmins IL-25, IL-33 and TSLP on eosinophilopoiesis is both indirect, since the secretion of IL-5 occurs by the ILC2, but it is also direct, since IL-33 can precede and promote IL-5 signaling in the eosinophil development process [39].
In addition the relevant role in promoting the proliferation, differentiation, and maturation of EoPs expressing IL-5Rα in the bone marrow, IL-5 is able to induce the release of eosinophils into the bloodstream, as well as the activation and survival of mature eosinophils at the tissue level, acting in combination with eotaxines [40].
The cytokines IL-3 and GM-CSF are also implicated in the activation and survival of tissue eosinophils through induction of Bcl-xL expression, but their action is less specific than that of IL-5 [41][42][43].
2.3. Eosinophil Degranulation
Once activated, eosinophils migrate from the peripheral blood to the inflammation site, in which they modulate the inflammatory response through the release of granules and therefore their contents [11][34]. Different degranulation processes are able to release specific granule contents (Figure 2):
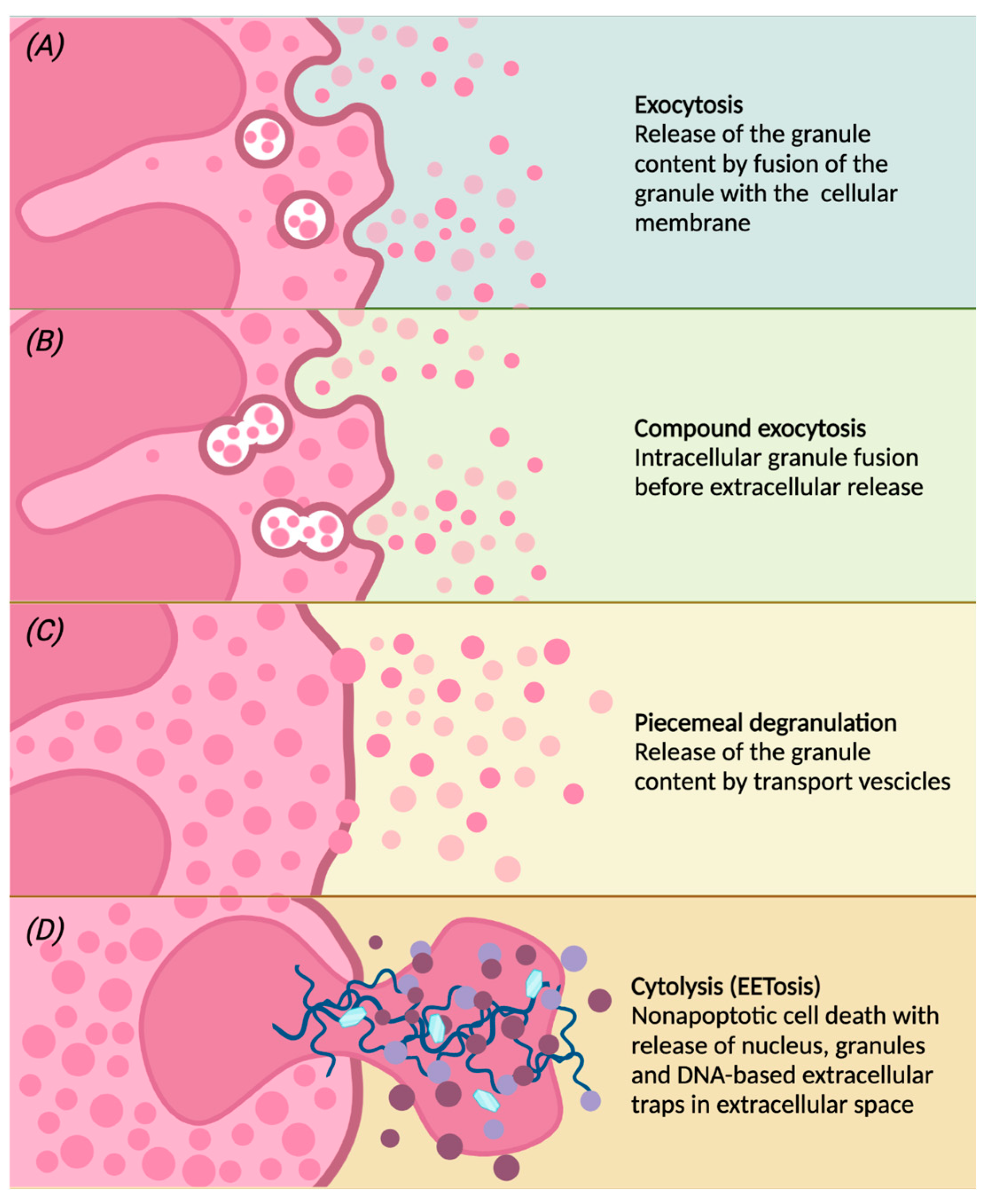
Figure 2. Following different stimulations, eosinophils can release the contents of the granules by classical exocytosis, compound exocytosis, piecemeal degranulation (PMD) or cytolysis. Conventional exocytosis consists of the release of granule content by fusion of the granule itself to the cellular membrane (panel A). Compound exocytosis is another type of exocytosis in which granules in which granules merge with each other before interacting with the cellular membrane (panel B). Piecemeal degranulation is the progressive and selective release of vesicles from specific granules and the unloading of their contents after the fusion with the cellular membrane (panel C). Cytolysis is a non-apoptotic form of cell death with rupture of the nuclear and plasma membrane, subsequent release of nuclear DNA and deposition of specific intact granules in the extracellular space. After cytolysis, there may be the release of eosinophilic extracellular traps (EETs), giving this peculiar form of cell death the characteristic name of EETosis (panel D).
(a) conventional exocytosis, in which the fusion of the granule directly with the cell membrane determines the release of the content of the specific granule itself;
(b) compound exocytosis, another type of exocytosis in which intracellular fusion of granules occurs prior to interaction with the plasma membrane and extracellular release;
(c) piecemeal degranulation (PMD), the most common mechanism of eosinophil degranulation, in which vesicles (that can be round or tubular) are released from specific granules and move towards the cell membrane to unload their content into the extracellular space [44]. Tubular vesicles tend to fold into a peculiar morphology and are therefore called “sombrero vesicles” [2];
(d) Cytolysis, a rapid non-apoptotic cell death in which the formation of vacuoles within cells occurs, with rupture of the nuclear and plasma membrane, the subsequent release of nuclear DNA and deposition of specific intact granules in the extracellular space. In this way, eosinophils release eosinophil extracellular traps (EETs), which consist of DNA fibers from the cellular nucleus [45]. Activated eosinophils are also capable of rapidly releasing other substances in addition to granule proteins into the extracellular space, such as bactericidal traps obtained from the combination of mitochondrial DNA and granule proteins; this type of cell death, which is characteristic of eosinophils, is also known as EETosis (“eosinophil extracellular trap cell death”) [15][46].
Finally, eosinophils can release exosomes into the extracellular environment; it has been demonstrated that eosinophils of asthmatic patients release greater amounts of extracellular vesicles (EVs) than those released by the eosinophils of healthy subjects [47]. EVs are important mediators produced by cellular processes [48]; this evidence strengthens the hypothesis that eosinophilic exosomes can be considered independent functional units, since, even in the absence of the cell of origin, they seem to be able to feed eosinophilic inflammation; however, exosomes and microvesicles are not the same entity: exosomes are generated by the fusion of multivesicular bodies (MVBs) with the plasma membrane, and microvesicles are shed by the outward vesiculation of the plasma membrane [49][50]. The pathogenetic relevance of eosinophil-derived EVs makes them a potential diagnostic and phenotypic biomarker of asthma, in particular of severe eosinophilic asthma [51][52].
3. Eosinophil Heterogeneity
Eosinophils were previously thought to be terminally differentiated cells upon their release from the bone marrow into the bloodstream, instead latest evidence demonstrated that eosinophils are able to further differentiate and mature in peripheral tissues, resulting in sub-populations with distinct phenotypic and functional profile [53].
Previous evidence from Mesnil et al. showed in mouse models a large population of eosinophils with a distinctive ring-shaped nucleus, both in absence of inflammation and following the development of dust-induced airway allergy, demonstrating the existence of lung resident eosinophils (rEosinophils) [54]; these cells, exclusively found in the lung parenchyma, express the surface receptors CD62L and CD125, intermediate levels of Siglec-F and low levels of CD101. Interestingly, even if they present the IL-5 surface receptor and react to IL-5 in vitro, rEosinophils seemed not to depend on IL-5 for their development [55], whereas the development of allergen-induced inflammatory eosinophils (iEosinophils) is known to be dependent on the activity of IL-5 [56] In addition, rEosinophils appear to have a more regulatory gene profile than iEosinophils, and mice without rheosinophils showed increased Th2 responses to inhaled allergens. Indeed, in allergic conditions, a large number of iEosinophils showing a segmented nucleus is recruited [56]. iEosinophils express low levels of CD62L, intermediate levels of CD125 and elevated levels of Siglec-F and CD101 on their surface, and are mainly concentrated in peribronchial areas [57].
Unlike mouse models, humans appear to have different eosinophil subsets based on cell density, in particular, normodense and hypodense eosinophils have been identified [58].
Normodense eosinophils from healthy individuals generally sediment at a density of 1.082 g/mL [58], while an increased number of hypodense eosinophils with a reduced density of <1.082 g/mL have been found in blood, BAL and lung tissues of patients with severe eosinophilic asthma [59][60]; these hypodense eosinophils were originally interpreted as activated eosinophils [61], since it has been shown that when normodense eosinophils are stimulated with GM-CSF, IL-3 or IL-5 and in the presence of fibroblasts, they switch to hypodense [62].
Hypodense eosinophils have been considered a “true” functional and phenotypic subset [63][64], since they highly react to activating stimuli. Indeed, after activation, hypodense eosinophils show greater survival, adhesion, oxygen metabolism, superoxide production, and antibody-dependent cytotoxicity than normodense [61].
Taken together, these studies have been useful in demonstrating that human lung eosinophils can be heterogeneous, however more studies are required for a better understanding of these subpopulations [55].
If the hypothesis of the existence of different subsets of human eosinophils is correct, it can significantly affect the choice of treatment in patients with severe eosinophilic asthma, between the total eradication of the eosinophilic lineage and the control of their IL-5-dependent development program [65].
4. Overview of Eosinophil-Driven Pathological Conditions
Elevated peripheral blood and tissue eosinophil counts can be found in several conditions, mainly allergic, rheumatological, infectious and neoplastic pathologies. Other possible conditions associated with eosinophilia have also been described, such as drug hypersensitivity, hematological and autoimmune diseases. Activation of eosinophils and subsequent release of eosinophilic mediators (mainly cytokines and type 2 chemokines) are potent pro-inflammatory effectors [66] and a significant association has been established between eosinophilia and some systemic inflammatory diseases [67]. The involvement of eosinophilia in inflammatory pathologies of the gastrointestinal, vascular, locomotor, and in general of the various mucous surfaces of the organism is known in the literature, and of course, eosinophils play key role in the pathogenesis of both upper and lower airway inflammation, with a spectrum of pathologic manifestations ranging from allergic rhinitis with or without nasal polyposis to asthma, and even allergic bronchopulmonary aspergillosis.
References
- Sastre, B.; Rodrigo-Muñoz, J.M.; Garcia-Sanchez, D.A.; Cañas, J.A.; Del Pozo, V. Eosinophils: Old Players in a New Game. J. Investig. Allergol. Clin. Immunol. 2018, 28, 289–304.
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4, 93.
- Gleich, G.J. Mechanisms of eosinophil-associated inflammation. J. Allergy Clin. Immunol. 2000, 105, 651–663.
- Michail, S.; Mezoff, E.; Abernathy, F. Role of Selectins in the Intestinal Epithelial Migration of Eosinophils. Pediatr. Res. 2005, 58, 644–647.
- Ying, S.; Meng, Q.; Zeibecoglou, K.; Robinson, D.S.; Macfarlane, A.; Humbert, M.; Kay, A.B. Eosinophil chemotactic chemokines (eotaxin, eotaxin-2, RANTES, monocyte chemoattractant protein-3 (MCP-3), and MCP-4), and C-C chemokine receptor 3 expression in bronchial biopsies from atopic and nonatopic (Intrinsic) asthmatics. J. Immunol. 1999, 163, 6321–6329.
- Stone, K.D.; Prussin, C.; Metcalfe, D.D. IgE, Mast Cells, Basophils, and Eosinophils. J. Allergy Clin. Immunol. 2010, 125, S73–S80.
- Muraki, M.; Gleich, G.J.; Kita, H. Antigen-Specific IgG and IgA, but Not IgE, Activate the Effector Functions of Eosinophils in the Presence of Antigen. Int. Arch. Allergy Immunol. 2011, 154, 119–127.
- Sedgwick, J.B.; Calhoun, W.J.; Vrtis, R.F.; Bates, M.E.; McAllister, P.K.; Busse, W.W. Comparison of airway and blood eosinophil function after in vivo antigen challenge. J. Immunol. 1992, 149, 3710–3718.
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820.
- Dembic, Z. The Function of Toll-like Receptors; Landes Bioscience: Austin, TX, USA, 2013.
- Kerr, S.C.; Gonzalez, J.R.; Schanin, J.; Peters, M.C.; Lambrecht, B.N.; Brock, E.C.; Charbit, A.; Ansel, K.M.; Youngblood, B.A.; Fahy, J.V. An anti-siglec-8 antibody depletes sputum eosinophils from asthmatic subjects and inhibits lung mast cells. Clin. Exp. Allergy 2020, 50, 904–914.
- Carr, T.F.; Berdnikovs, S.; Simon, H.-U.; Bochner, B.S.; Rosenwasser, L.J. Eosinophilic bioactivities in severe asthma. World Allergy Organ. J. 2016, 9, 21.
- Kiwamoto, T.; Kawasaki, N.; Paulson, J.C.; Bochner, B.S. Siglec-8 as a drugable target to treat eosinophil and mast cell-associated conditions. Pharmacol. Ther. 2012, 135, 327–336.
- Mori, Y.; Iwasaki, H.; Kohno, K.; Yoshimoto, G.; Kikushige, Y.; Okeda, A.; Uike, N.; Niiro, H.; Takenaka, K.; Nagafuji, K.; et al. Identification of the human eosinophil lineage-committed progenitor: Revision of phenotypic definition of the human common myeloid progenitor. J. Exp. Med. 2009, 206, 183–193.
- Klion, A. Recent advances in understanding eosinophil biology. F1000Research 2017, 6, 1084.
- Dorman, S.C.; Efthimiadis, A.; Babirad, I.; Watson, R.M.; Denburg, J.A.; Hargreave, F.E.; O’Byrne, P.M.; Sehmi, R. Sputum CD34+IL-5Ralpha+ cells increase after allergen: Evidence for in situ eosinophilopoiesis. Am. J. Respir. Crit. Care Med. 2004, 169, 573–577.
- Rådinger, M.; Bossios, A.; Sjöstrand, M.; Lu, Y.; Malmhäll, C.; Dahlborn, A.-K.; Lee, J.J.; Lötvall, J. Local proliferation and mobilization of CCR3(+) CD34(+) eosinophil-lineage-committed cells in the lung. Immunology 2011, 132, 144–154.
- Sehmi, R.; Howie, K.; Sutherland, D.R.; Schragge, W.; O’Byrne, P.M.; Denburg, J.A. Increased levels of CD34+ hemopoietic progenitor cells in atopic subjects. Am. J. Respir. Cell Mol. Biol. 1996, 15, 645–655.
- Sehmi, R.; Smith, S.G.; Kjarsgaard, M.; Radford, K.; Boulet, L.-P.; Lemiere, C.; Prazma, C.M.; Ortega, H.; Martin, J.G.; Nair, P. Role of local eosinophilopoietic processes in the development of airway eosinophilia in prednisone-dependent severe asthma. Clin. Exp. Allergy 2016, 46, 793–802.
- Du, J.; Stankiewicz, M.J.; Liu, Y.; Xi, Q.; Schmitz, J.E.; Lekstrom-Himes, J.A.; Ackerman, S.J. Novel Combinatorial Interactions of GATA-1, PU.1, and C/EBPε Isoforms Regulate Transcription of the Gene Encoding Eosinophil Granule Major Basic Protein. J. Biol. Chem. 2002, 277, 43481–43494.
- Yu, C.; Cantor, A.B.; Yang, H.; Browne, C.; Wells, R.A.; Fujiwara, Y.; Orkin, S.H. Targeted Deletion of a High-Affinity GATA-binding Site in the GATA-1 Promoter Leads to Selective Loss of the Eosinophil Lineage In Vivo. J. Exp. Med. 2002, 195, 1387–1395.
- Dougan, M.; Dranoff, G.; Dougan, S.K. GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 2019, 50, 796–811.
- Wechsler, M.E.; Munitz, A.; Ackerman, S.J.; Drake, M.G.; Jackson, D.J.; Wardlaw, A.J.; Dougan, S.K.; Berdnikovs, S.; Schleich, F.; Matucci, A.; et al. Eosinophils in Health and Disease: A State-of-the-Art Review. Mayo Clin. Proc. 2021, 96, 2694–2707.
- Eng, S.S.; DeFelice, M.L. The Role and Immunobiology of Eosinophils in the Respiratory System: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 50, 140–158.
- Bochner, B.S. The eosinophil. Ann. Allergy. Asthma. Immunol. 2018, 121, 150–155.
- Ramirez, G.A.; Yacoub, M.-R.; Ripa, M.; Mannina, D.; Cariddi, A.; Saporiti, N.; Ciceri, F.; Castagna, A.; Colombo, G.; Dagna, L. Eosinophils from Physiology to Disease: A Comprehensive Review. BioMed Res. Int. 2018, 2018, 9095275.
- Katsoulis, K.; Kipourou, M.; Loukides, S. Reduction/elimination of blood eosinophils in severe asthma: Should there be a safety consideration? Expert Opin. Biol. Ther. 2021, 22, 377–384.
- Rosenberg, H.F.; Phipps, S.; Foster, P.S. Eosinophil trafficking in allergy and asthma. J. Allergy Clin. Immunol. 2007, 119, 1303–1310; quiz 1311–1312.
- Woltmann, G.; McNulty, C.A.; Dewson, G.; Symon, F.A.; Wardlaw, A.J. Interleukin-13 induces PSGL-1/P-selectin-dependent adhesion of eosinophils, but not neutrophils, to human umbilical vein endothelial cells under flow. Blood 2000, 95, 3146–3152.
- Janson, C.; Bjermer, L.; Lehtimäki, L.; Kankaanranta, H.; Karjalainen, J.; Altraja, A.; Yasinska, V.; Aarli, B.; Rådinger, M.; Hellgren, J.; et al. Eosinophilic airway diseases: Basic science, clinical manifestations and future challenges. Eur. Clin. Respir. J. 2022, 9, 2040707.
- Mjösberg, J.; Spits, H. Human innate lymphoid cells. J. Allergy Clin. Immunol. 2016, 138, 1265–1276.
- Boberg, E.; Johansson, K.; Malmhäll, C.; Calvén, J.; Weidner, J.; Rådinger, M. Interplay Between the IL-33/ST2 Axis and Bone Marrow ILC2s in Protease Allergen-Induced IL-5-Dependent Eosinophilia. Front. Immunol. 2020, 11, 1058.
- Johansson, K.; Malmhäll, C.; Ramos-Ramírez, P.; Rådinger, M. Bone marrow type 2 innate lymphoid cells: A local source of interleukin-5 in interleukin-33-driven eosinophilia. Immunology 2018, 153, 268–278.
- Wardlaw, A.J. Molecular basis for selective eosinophil trafficking in asthma: A multistep paradigm. J. Allergy Clin. Immunol. 1999, 104, 917–926.
- Venge, J.; Lampinen, M.; Håkansson, L.; Rak, S.; Venge, P. Identification of IL-5 and RANTES as the major eosinophil chemoattractants in the asthmatic lung. J. Allergy Clin. Immunol. 1996, 97, 1110–1115.
- Sjöberg, L.C.; Nilsson, A.Z.; Lei, Y.; Gregory, J.A.; Adner, M.; Nilsson, G.P. Interleukin 33 exacerbates antigen driven airway hyperresponsiveness, inflammation and remodeling in a mouse model of asthma. Sci. Rep. 2017, 7, 4219.
- Watson, B.; Gauvreau, G.M. Thymic stromal lymphopoietin: A central regulator of allergic asthma. Expert Opin. Ther. Targets 2014, 18, 771–785.
- Yao, X.; Sun, Y.; Wang, W.; Sun, Y. Interleukin (IL)-25: Pleiotropic roles in asthma. Respirology 2016, 21, 638–647.
- Chen, R.; Smith, S.G.; Salter, B.; El-Gammal, A.; Oliveria, J.P.; Obminski, C.; Watson, R.; O’Byrne, P.M.; Gauvreau, G.M.; Sehmi, R. Allergen-induced Increases in Sputum Levels of Group 2 Innate Lymphoid Cells in Subjects with Asthma. Am. J. Respir. Crit. Care Med. 2017, 196, 700–712.
- Johnston, L.K.; Hsu, C.-L.; Krier-Burris, R.A.; Chhiba, K.D.; Chien, K.B.; McKenzie, A.; Berdnikovs, S.; Bryce, P.J. IL-33 Precedes IL-5 in Regulating Eosinophil Commitment and Is Required for Eosinophil Homeostasis. J. Immunol. 2016, 197, 3445–3453.
- Palframan, R.T.; Collins, P.D.; Williams, T.J.; Rankin, S.M. Eotaxin Induces a Rapid Release of Eosinophils and Their Progenitors From the Bone Marrow. Blood 1998, 91, 2240–2248.
- Rodrigo-Muñoz, J.M.; Gil-Martínez, M.; Sastre, B.; del Pozo, V. Emerging Evidence for Pleiotropism of Eosinophils. Int. J. Mol. Sci. 2021, 22, 7075.
- Esnault, S.; Kelly, E.A. Essential Mechanisms of Differential Activation of Eosinophils by IL-3 Compared to GM-CSF and IL-5. Crit. Rev. Immunol. 2016, 36, 429–444.
- Schwartz, C.; Willebrand, R.; Huber, S.; Rupec, R.A.; Wu, D.; Locksley, R.; Voehringer, D. Eosinophil-specific deletion of IκBα in mice reveals a critical role of NF-κB-induced Bcl-xL for inhibition of apoptosis. Blood 2015, 125, 3896–3904.
- Fettrelet, T.; Gigon, L.; Karaulov, A.; Yousefi, S.; Simon, H.-U. The Enigma of Eosinophil Degranulation. Int. J. Mol. Sci. 2021, 22, 7091.
- Ueki, S.; Tokunaga, T.; Melo, R.C.N.; Saito, H.; Honda, K.; Fukuchi, M.; Konno, Y.; Takeda, M.; Yamamoto, Y.; Hirokawa, M.; et al. Charcot-Leyden crystal formation is closely associated with eosinophil extracellular trap cell death. Blood 2018, 132, 2183–2187.
- Yousefi, S.; Gold, J.A.; Andina, N.; Lee, J.J.; Kelly, A.M.; Kozlowski, E.; Schmid, I.; Straumann, A.; Reichenbach, J.; Gleich, G.J.; et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008, 14, 949–953.
- Mazzeo, C.; Cañas, J.A.; Zafra, M.P.; Rojas Marco, A.; Fernández-Nieto, M.; Sanz, V.; Mittelbrunn, M.; Izquierdo, M.; Baixaulli, F.; Sastre, J.; et al. Exosome secretion by eosinophils: A possible role in asthma pathogenesis. J. Allergy Clin. Immunol. 2015, 135, 1603–1613.
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Raposo, G.; Stahl, P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510.
- Choi, Y.; Sim, S.; Park, H.-S. Distinct functions of eosinophils in severe asthma with type 2 phenotype: Clinical implications. Korean J. Intern. Med. 2020, 35, 823–833.
- Sastre, B.; Cañas, J.A.; Rodrigo-Muñoz, J.M.; Del Pozo, V. Novel Modulators of Asthma and Allergy: Exosomes and MicroRNAs. Front. Immunol. 2017, 8, 826.
- Van Hulst, G.; Batugedara, H.M.; Jorssen, J.; Louis, R.; Bureau, F.; Desmet, C.J. Eosinophil diversity in asthma. Biochem. Pharmacol. 2020, 179, 113963.
- Mesnil, C.; Raulier, S.; Paulissen, G.; Xiao, X.; Birrell, M.A.; Pirottin, D.; Janss, T.; Starkl, P.; Ramery, E.; Henket, M.; et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Investig. 2016, 126, 3279–3295.
- Davoine, F.; Lacy, P. Eosinophil cytokines, chemokines, and growth factors: Emerging roles in immunity. Front. Immunol. 2014, 5, 570.
- Yukawa, T.; Kroegel, C.; Evans, P.; Fukuda, T.; Chung, K.F.; Barnes, P.J. Density heterogeneity of eosinophil leucocytes: Induction of hypodense eosinophils by platelet-activating factor. Immunology 1989, 68, 140–143.
- Frick, W.E.; Sedgwick, J.B.; Busse, W.W. The appearance of hypodense eosinophils in antigen-dependent late phase asthma. Am. Rev. Respir. Dis. 1989, 139, 1401–1406.
- Fukuda, T.; Dunnette, S.L.; Reed, C.E.; Ackerman, S.J.; Peters, M.S.; Gleich, G.J. Increased numbers of hypodense eosinophils in the blood of patients with bronchial asthma. Am. Rev. Respir. Dis. 1985, 132, 981–985.
- Fukuda, T.; Gleich, G.J. Heterogeneity of human eosinophils. J. Allergy Clin. Immunol. 1989, 83, 369–373.
- Campbell, H.D.; Tucker, W.Q.; Hort, Y.; Martinson, M.E.; Mayo, G.; Clutterbuck, E.J.; Sanderson, C.J.; Young, I.G. Molecular cloning, nucleotide sequence, and expression of the gene encoding human eosinophil differentiation factor (interleukin 5). Proc. Natl. Acad. Sci. USA 1987, 84, 6629–6633.
- Garcia-Romo, G.S.; Caielli, S.; Vega, B.; Connolly, J.; Allantaz, F.; Xu, Z.; Punaro, M.; Baisch, J.; Guiducci, C.; Coffman, R.L.; et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 2011, 3, 73ra20.
- Chatfield, S.M.; Thieblemont, N.; Witko-Sarsat, V. Expanding Neutrophil Horizons: New Concepts in Inflammation. J. Innate Immun. 2018, 10, 422–431.
- Laviolette, M.; Gossage, D.L.; Gauvreau, G.; Leigh, R.; Olivenstein, R.; Katial, R.; Busse, W.W.; Wenzel, S.; Wu, Y.; Datta, V.; et al. Effects of benralizumab on airway eosinophils in asthmatic patients with sputum eosinophilia. J. Allergy Clin. Immunol. 2013, 132, 1086–1096.e5.
- Kanda, A.; Yun, Y.; Bui, D.V.; Nguyen, L.M.; Kobayashi, Y.; Suzuki, K.; Mitani, A.; Sawada, S.; Hamada, S.; Asako, M.; et al. The multiple functions and subpopulations of eosinophils in tissues under steady-state and pathological conditions. Allergol. Int. Off. J. Jpn; thusc. Allergol. 2021, 70, 9–18.
- Lombardi, C.; Passalacqua, G. Eosinophilia and diseases: Clinical revision of 1862 cases. Arch. Intern. Med. 2003, 163, 1371–1373.
- Lombardi, C.; Berti, A.; Cottini, M. The emerging roles of eosinophils: Implications for the targeted treatment of eosinophilic-associated inflammatory conditions. Curr. Res. Immunol. 2022, 3, 42–53.
More
Information
Subjects:
Cell Biology; Allergy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
9.5K
Revisions:
3 times
(View History)
Update Date:
09 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





