
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Makoto Tsuneoka | + 2311 word(s) | 2311 | 2020-10-13 08:00:45 | | | |
| 2 | Camila Xu | Meta information modification | 2311 | 2020-10-20 11:26:49 | | | | |
| 3 | Camila Xu | Meta information modification | 2311 | 2020-10-26 09:31:50 | | |
Video Upload Options
KDM2A is a member of one group of α-ketoglutarate-dependent oxygenases. KDM2A in the rDNA promoter is activated by nutrient starvation, to reduce rRNA transcription and cell proliferation. While gallic acid functions as an antioxidant, gallic acid autoxidation also produces significant levels of reactive oxygen species (ROS). Gallic acid activates KDM2A to reduce rRNA transcription and cell proliferation in breast cancer MCF-7 cells but not in non-tumorous MCS10A cells. The activation of KDM2A by gallic acid depends on ROS production and AMPK activation.
1. Introduction
The ribosome is a unique machine for protein synthesis in organisms, and the number of ribosomes widely affects cellular activities [1][2]. Ribosome biogenesis consumes cellular materials supplied by nutrients and their metabolites, and the level of ribosome biogenesis reflects intracellular conditions. Therefore, cells should have mechanisms to control ribosome production by detecting intracellular conditions. The control of rRNA transcription is a major factor determining the amount of ribosome production [3], and thus, the levels of rRNA transcription affect multiple cellular activities including cell proliferation [4][5].The control of chromatin structure affects transcription and involves chemical modification of DNA and histones including the addition or removal of methyl groups [7][8]. Lysine-specific histone demethylase (KDM) consists of two main classes: a flavin adenine dinucleotide-dependent amine oxidase, and an Fe(II) and α-ketoglutarate-dependent hydroxylase that has the JmjC domain as an active center of the enzyme. The JmjC-type KDMs are one group of α-ketoglutarate-dependent oxygenases that catalyze a remarkably diverse range of oxidative reactions [9]. The JmjC-type KDMs remove methyl groups on lysine in the specific amino acid stretches of histones, and the selectivity of substrates is determined by the structure of the JmjC domain and also the dynamics of the amino acid sequence near the JmjC domain [10][11]. KDMs also distinguish the number of methyl groups attached to lysine [7]. Dysregulation of JmjC-type KDM family members is implicated in many diseases, including developmental disorders and cancers. Recently, increasing numbers of studies have suggested that epigenetic regulators control rRNA transcription [3][4]. We previously showed that a JmjC-type KDM, KDM2A, decreases the dimethylated lysine 36 of histone H3 (H3K36me2) but not trimethylated lysine 36 of H3 (H3K36me3) in the rRNA gene (rDNA) promoter, and represses rRNA transcription under starvation in breast cancer cells [12][13]. Further, we found that glucose depletion activates AMP-activated protein kinase (AMPK), which is required for KDM2A to reduce rRNA transcription and cell proliferation of breast cancer cells [12]. We also found that metformin, known as a treatment for type 2 diabetes, activates KDM2A to reduce rRNA transcription and cell proliferation [14]. A 4-h treatment with metformin not only activated AMPK, but also independently decreased the intracellular succinate level [14]. The activation of KDM2A by metformin required both AMPK activity and a decrease of the intracellular succinate level [14], suggesting that reduction of intracellular succinate, in addition to AMPK activation, is a mechanism by which KDM2A is activated to reduce rRNA transcription.
Gallic acid [3,4,5-trihydroxybenzoic acid], a natural botanic phenolic compound, is abundantly found in green tea, grapes, and red wine [15][16][17]. Gallic acid functions as an antioxidant to exert a wide range of pharmacological properties including anti-obesity, anti-inflammation, and anti-cancer activities [18]. On the other hand, reactive oxygen species (ROS) was also generated when gallic acid was added to the cell culture medium [19]. Gallic acid autoxidation produced significant levels of H2O2 and O2 • − (superoxide), and the number of apoptotic cells increased depending on the gallic acid concentration. Intracellular ROS levels that increased by gallic acid were reduced by N-acetyl-l-cysteine (NAC) and l-glutathione (GSH) [20]. Therefore, gallic acid can produce ROS in addition to its antioxidant activity.
Because KDM2A has the activity to reduce rRNA transcription and proliferation of breast cancer cells [12], we previously proposed that the reduction of rRNA transcription by KDM2A specifically in cancer cells may be applied to the treatment of breast cancers [21].
2. Gallic Acid Induced KDM2A Dependent-Reduction of rRNA Transcription and Cell Proliferation in MCF-7 Cells
A food-additive compounds library (Sigma; #S990043-FDS1) was used to screen compounds that control cell proliferation of breast cancer cell line MCF-7 cells. Cells were treated with 133 μM of each compound for 2 days, and cell numbers were detected with CyQUANT Direct Cell Proliferation Assay by measuring the amounts of DNA. Nine out of 77 compounds decreased cell numbers to less than 80%, and 15 compounds tended to increase cell numbers. Among the compounds, gallic acid reduced cell numbers in a KDM2A-dependent manner. Therefore, we focused our study on the mechanism of gallic acid to decrease cell proliferation of MCF-7 cells. When MCF-7 cells were treated with various concentrations of gallic acid for 2 days, treatment with 50 or 200 μM gallic acid decreased the cell number, but 12.5 μM gallic acid did not (Figure 1A). The KDM2A knockdown significantly alleviated the decrease of cell number by 50 μM gallic acid, but not that by 200 μM gallic acid (Figure 1A). These results suggested that the treatment with 50 μM gallic acid decreased cell proliferation through KDM2A.
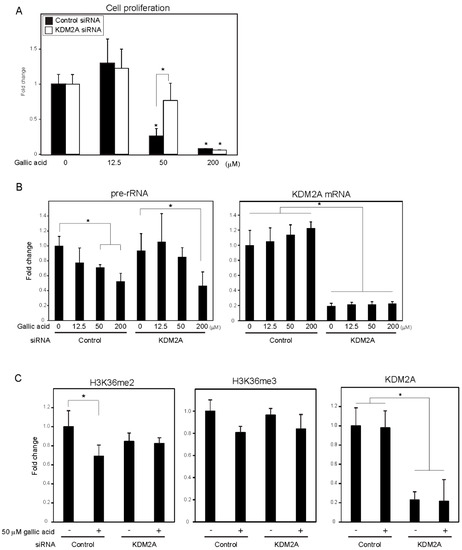
Figure 1. Gallic acid induced KDM2A-dependent reductions of cell proliferation, rRNA transcription, and H3K36me2 in rDNA promoter in MCF-7 cells. (A) Gallic acid decreased proliferation of MCF-7 cells through KDM2A. MCF-7 cells transfected with control siRNA or siRNA for KDM2A were cultured with gallic acid as indicated concentrations for 2 days. Cell numbers were measured by CyQUANT® Direct Cell Proliferation Assay detecting DNA content. (B) The treatment with 50 μM gallic acid reduces rRNA transcription through KDM2A. MCF-7 cells transfected with control siRNA or siRNA for KDM2A were treated with gallic acid at indicated concentrations for 4 h. Total RNAs were isolated and analyzed by quantitative real-time PCR (qRT-PCR) to detect rRNA transcription (pre-rRNA) (left panel) and KDM2A mRNA (right panel). The ratios of the values for cells treated with various conditions to those for cells treated with control siRNA without gallic acid are shown. (C) KDM2A-dependent reduction of H3K36me2 marks in rDNA promoter by treatment with 50 μM gallic acid. MCF-7 cells transfected with control siRNA or siRNA for KDM2A were treated with 50 μM gallic acid for 4 h. The levels of H3K36me2, H3K36me3, and KDM2A in the rDNA promoter were analyzed by ChIP assay. The results are expressed as fold changes of the values under various conditions to those in cells cultured with control siRNA without gallic acid treatment. All experiments were performed more than three times, and the mean values with standard deviations are indicated. * p < 0.05.
To investigate whether the decrease of cell numbers by gallic acid was associated with the decrease of rRNA transcription, the levels of rRNA transcription were measured at 4 h after gallic acid treatment. The treatment of cells with gallic acid decreased rRNA transcription in a dose-dependent manner (Figure 1B), and the KDM2A knockdown alleviated the decrease of rRNA transcription in cells treated with 50 μM gallic acid (Figure 1B). In the case of 200 μM gallic acid, the levels of rRNA transcription were reduced even when KDM2A was knocked down (Figure 1B). Treatment with 50 μM gallic acid decreased the level of H3K36me2, a direct substrate of KDM2A, in the rDNA promoter, depending on KDM2A (Figure 1C), but did not significantly affect the levels of neither KDM2A nor H3K36me3 in the rDNA promoter (Figure 1C). The demethylation of JmjC-type enzymes proceeded by a side reaction that produced succinate from α-ketoglutarate (α-KG) [24], and it was shown that succinate can inhibit the demethylase activity of KDM2A [12][13][14]. The addition of a cell-permeable succinate, dimethyl succinate (DMS), to the medium inhibited the reductions of H3K36me2 in the rDNA promoter and rRNA transcription induced by 50 μM gallic acid. These results suggest that 50 μM gallic acid activated the demethylase activity of KDM2A to reduce rRNA transcription and cell proliferation.
3. Gallic Acid Elevated ROS Production and AMPK Activation, both of which are Required for KDM2A to Regulate H3K36me2 Levels in the rDNA Promoter and rRNA Transcription
It was reported that gallic acid showed anti-cancer activity in some cancer cells that probably involved the production of ROS [25][26]. We measured the levels of intracellular ROS using 2′,7′-dichlorofluorescein (DCF) diacetate, a cell-permeable probe. It was found that treatment with 50 μM gallic acid increased the DCF signal (Figure 3A). Antioxidants, such as N-acetylcysteine (NAC) and glutathione (GSH), reduced the DCF signal increased by 50 μM gallic acid (Figure 3A). These results show that gallic acid treatment increased the level of intracellular ROS in MCF-7 cells. The NAC and GSH treatments impaired the reduction of rRNA transcription (Figure 3B) and H3K36me2 marks in the rDNA promoter (Figure 3C) induced by 50 μM gallic acid. The levels of H3K36me3 and KDM2A in the rDNA promoter were not significantly changed under these conditions (Figure 3C). The results indicate that the increase of ROS by gallic acid is required for the induction of KDM2A activity to reduce rRNA transcription.
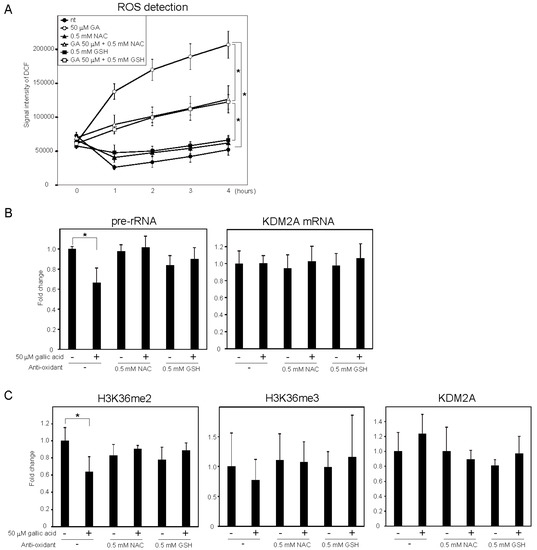
Figure 2. ROS production by gallic acid was required for the repression of rRNA transcription mediated by KDM2A in MCF-7 cells. (A) Gallic acid increases ROS production in MCF-7 cells. MCF-7 cells cultured with cell-permeable ROS probe DCFDA were cultured with or without 50 μM gallic acid (GA) in the presence or absence of 0.5 mM N-acetyl-l-cysteine (NAC) or l-glutathione (GSH). At the indicated times, the increases in the signal intensities of DCF by ROS in cells were measured. (B) Effects of anti-oxidants, NAC and GSH, on the reduction of rRNA transcription by gallic acid. MCF-7 cells treated with or without 50 μM gallic acid in the presence or absence of 0.5 mM NAC or 0.5 mM GSH for 4 h. Total RNAs were isolated and analyzed by qRT-PCR to detect pre-rRNA (left panel) and KDM2A mRNA (right panel). The ratios of the values for cells treated with various conditions to those for cells treated without gallic acid and anti-oxidants are shown. (C) Effects of anti-oxidants, NAC and GSH, on the reduction of H3K36me2 marks in the rDNA promoter by gallic acid. MCF-7 cells were cultured for 4 h with or without 50 μM gallic acid in the presence or absence of NAC or GSH. The levels of H3K36me2, H3K36me3, and KDM2A in the rDNA promoter were analyzed by ChIP assays. The results are expressed as fold changes to the values in various conditions to those without gallic acid and anti-oxidants. All experiments were performed more than three times, and the mean values with standard deviations are indicated. * p < 0.05.
Next, whether the oxidative stress alone repressed rRNA transcription through KDM2A was tested. When cells were treated with various concentrations of H2O2, rRNA transcription was reduced and the KDM2A knockdown slightly alleviated the reduction of rRNA transcription at 12.5 μM H2O2. However, the level of H3K36me2 in the rDNA promoter was not reduced by 12.5 μM H2O2. Therefore, H2O2 alone did not activate the KDM2A demethylase activity in the rDNA promoter.
Previously, we showed that AMPK activity was required for KDM2A to reduce the levels of H3K36me2 in the rDNA promoter and rRNA transcription under glucose starvation [12] or by metformin [14]. Treatment with gallic acid was reported to activate AMPK in the liver cancer cell line HepG2 cells [27]. When MCF-7 cells were treated with 0, 12.5, 50, or 200 μM gallic acid for 4 h, the level of Thr172 phosphorylated AMPKα, the activation mark of AMPK, was increased (Figure 3A, pAMPK). The amounts of total AMPKα (tAMPK) and β-actin were not changed under these conditions (Figure 3A, tAMPK), showing that the treatment with gallic acid activated AMPK. Treatments with compound C or SBI-0206965, known as inhibitors of AMPK, impaired the 50 μM gallic acid-dependent reductions of rRNA transcription (Figure 3B) and H3K36me2 in the rDNA promoter (Figure 3C). These results suggest that the activation of AMPK by 50 μM gallic acid is required for the reduction of rRNA transcription through KDM2A. The treatments with NAC and GSH did not impair AMPK activation by 50 μM gallic acid, showing that gallic acid causes the AMPK activation without elevating ROS production. Together, these results suggest that gallic acid elevates AMPK activation and ROS production, both of which are required for KDM2A to regulate rRNA transcription through its histone demethylase activity in the rDNA promoter.
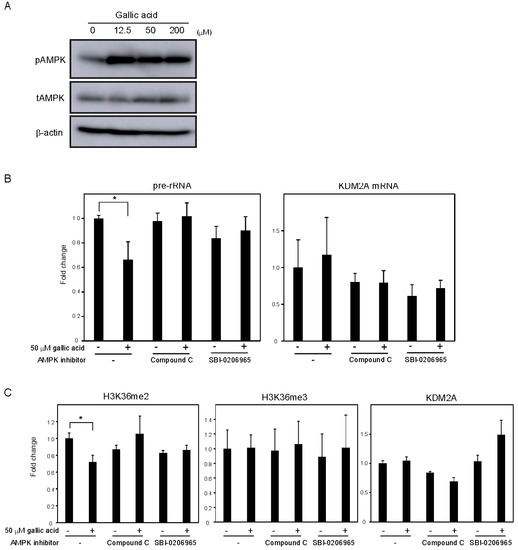
Figure 3. AMPK activation is required for the KDM2A-mediated repression of rRNA transcription by gallic acid in MCF-7 cells. (A) Activation of AMPK by gallic acid in MCF-7 cells. MCF-7 cells were treated with gallic acid as indicated concentrations for 4 h. Cells were lysed and analyzed for the levels of phosphorylated-AMPKα (Thr172) (pAMPK), total AMPKα (tAMPK), and β-actin by immunoblotting. (B) Requirement of AMPK activity for the reduction of rRNA transcription by gallic acid. MCF-7 cells treated with or without 50 μM gallic acid in the presence or absence of AMPK inhibitor, 10 μM compound C or 5 μM SBI-0206965, for 4 h. Total RNAs were isolated and analyzed by qRT-PCR to detect pre-rRNA (left panel) and KDM2A mRNA (right panel). The ratios of the values for cells treated with various conditions to those for cells treated without gallic acid and AMPK inhibitors are shown. The experiments were performed three times (n = 3), and the mean values with standard deviations are indicated. * p < 0.05. (C) Requirement of AMPK activity for the reduction of H3K36me2 mark in the rDNA promoter by gallic acid. MCF-7 cells treated with or without 50 μM gallic acid in the presence of 10 μM compound C or 5 μM SBI-0206965 for 4 h. The levels of H3K36me2, H3K36me3, and KDM2A in the rDNA promoter were analyzed by ChIP assay. The results are expressed as fold changes to the values in various conditions to those without gallic acid and AMPK inhibitors. The experiments were performed three times (n = 3), and the mean values with standard deviations are indicated. * p < 0.05.
References
- Grummt, I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003, 17, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.O.J. (Ed.) The Nucleolus; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Kusnadi, E.P.; Hannan, K.M.; Hicks, R.J.; Hannan, R.D.; Pearson, R.B.; Kang, J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 2015, 556, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, S.; Bierhoff, H. Regulation of RNA polymerase I transcription in development, disease, and aging. Annu. Rev. Biochem. 2018, 87, 51–73. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.T.M. Gene Expression and Regulation in Mammalian Cells; Uchiumi, F., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Ferreira, R.; Schneekloth, J.S.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells 2020, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef]
- Cortopassi, W.A.; Simion, R.; Honsby, C.E.; França, T.C.; Paton, R.S. Dioxygen binding in the active site of histone demethylase JMJD2A and the role of the protein environment. Chemistry 2015, 21, 18983–18992. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Ramanan, R.; Waheed, S.O.; Ainsley, J.; Evison, M.; Ames, J.M.; Schofield, C.J.; Karabencheva-Christova, T.G.; Christov, C.Z. Conformational dynamics underlies different functions of human KDM7 histone demethylases. Chemistry 2019, 25, 5422–5426. [Google Scholar] [CrossRef]
- Tanaka, Y.; Yano, H.; Ogasawara, S.; Yoshioka, S.; Imamura, H.; Okamoto, K.; Tsuneoka, M. Mild glucose starvation induces KDM2A-mediated H3K36me2 demethylation through AMPK to reduce rRNA transcription and cell proliferation. Mol. Cell Biol. 2015, 35, 4170–4184. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Okamoto, K.; Teye, K.; Umata, T.; Yamagiwa, N.; Suto, Y.; Zhang, Y.; Tsuneoka, M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010, 29, 1510–1522. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Konishi, A.; Obinata, H.; Tsuneoka, M. Metformin activates KDM2A to reduce rRNA transcription and cell proliferation by dual regulation of AMPK activity and intracellular succinate level. Sci. Rep. 2019, 9, 18694. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L. Naturally occurring food toxicants: Phenolic substances of plant origin common in foods. Adv. Food Res. 1981, 27, 149–242. [Google Scholar] [CrossRef] [PubMed]
- Anantharaju, P.G.; Gowda, P.C.; Vimalambike, M.G.; Madhunapantula, S.V. An overview on the role of dietary phenolics for the treatment of cancers. Nutr. J. 2016, 15, 99. [Google Scholar] [CrossRef] [PubMed]
- Gallic Acid. National Library of Medicine National Center for Biotechnology Information. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Gallic_acid (accessed on 3 October 2020).
- Aggarwal, V.; Tuli, H.S.; Tania, M.; Srivastava, S.; Ritzer, E.E.; Pandey, A.; Aggarwal, D.; Barwal, T.S.; Jain, A.; Kaur, G.; et al. Molecular mechanisms of action of epigallocatechin gallate in cancer: Recent trends and advancement. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Grzesik, M.; Bartosz, G.; Stefaniuk, I.; Pichla, M.; Namiesnik, J.; Sadowska-Bartosz, I. Dietary antioxidants as a source of hydrogen peroxide. Food Chem. 2019, 278, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.H.; Mazzio, E.; Badisa, R.B.; Zhu, Z.P.; Agharahimi, M.; Oriaku, E.T.; Goodman, C.B. Autoxidation of gallic acid induces ROS-dependent death in human prostate cancer LNCaP cells. Anticancer Res. 2012, 32, 1595–1602. [Google Scholar] [PubMed]
- Okamoto, K.; Tanaka, Y.; Ogasawara, S.; Obuse, C.; Nakayama, J.I.; Yano, H.; Tsuneoka, M. KDM2A-dependent reduction of rRNA transcription on glucose starvation requires HP1 in cells, including triple-negative breast cancer cells. Oncotarget 2019, 10, 4743–4760. [Google Scholar] [CrossRef]
- Nelson, J.D.; Denisenko, O.; Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006, 1, 179–185. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Marengo, B.; Zuccari, G.; Turrini, F.; Domenicotti, C. Dendrimer nanodevices and gallic acid as novel strategies to fight chemoresistance in neuroblastoma cells. Nanomaterials 2020, 10, 1243. [Google Scholar] [CrossRef] [PubMed]
- Tor, Y.S.; Yazan, L.S.; Foo, J.B.; Wibowo, A.; Ismail, N.; Cheah, Y.K.; Abdullah, R.; Ismail, M.; Ismail, I.S.; Yeap, S.K. Induction of apoptosis in MCF-7 cells via oxidative stress generation, mitochondria-dependent and caspase-independent pathway by ethyl acetate extract of dillenia suffruticosa and its chemical profile. PLoS ONE 2015, 10, e0127441. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Sato, A.; Kishimoto, Y.; Mabashi-Asazuma, H.; Kondo, K.; Iida, K. Gallic acid inhibits lipid accumulation via AMPK pathway and suppresses apoptosis and macrophage-mediated inflammation in hepatocytes. Nutrients 2020, 12, 1479.




