Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mario I Vega | -- | 3682 | 2022-08-15 17:53:01 | | | |
| 2 | Vivi Li | -1 word(s) | 3681 | 2022-08-16 04:50:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Morales-Martínez, M.; Vega, M.I. MicroRNA-7 (MiR-7) in Cancer Physiopathology. Encyclopedia. Available online: https://encyclopedia.pub/entry/26157 (accessed on 08 February 2026).
Morales-Martínez M, Vega MI. MicroRNA-7 (MiR-7) in Cancer Physiopathology. Encyclopedia. Available at: https://encyclopedia.pub/entry/26157. Accessed February 08, 2026.
Morales-Martínez, Mario, Mario I. Vega. "MicroRNA-7 (MiR-7) in Cancer Physiopathology" Encyclopedia, https://encyclopedia.pub/entry/26157 (accessed February 08, 2026).
Morales-Martínez, M., & Vega, M.I. (2022, August 15). MicroRNA-7 (MiR-7) in Cancer Physiopathology. In Encyclopedia. https://encyclopedia.pub/entry/26157
Morales-Martínez, Mario and Mario I. Vega. "MicroRNA-7 (MiR-7) in Cancer Physiopathology." Encyclopedia. Web. 15 August, 2022.
Copy Citation
miRNAs are non-coding RNA sequences of approximately 22 nucleotides that interact with genes by inhibiting their translation through binding to their 3′ or 5′ UTR regions. Following their discovery, the role they play in the development of various pathologies, particularly cancer, has been studied. In this context, miR-7 is described as an important factor in the development of cancer because of its role as a tumor suppressor, regulating a large number of genes involved in the development and progression of cancer. Data support the function of miR-7 as a prognostic biomarker in cancer, and miR-7 has been proposed as a strategy in cancer therapy.
miR-7
cancer
cancer progression
clinical implication
gene expression
1. Introduction
Cancer is defined by the WHO as a broad group of diseases that can affect any part of the body, characterized by the rapid proliferation of abnormal cells that extend beyond their usual limits and can invade adjacent parts of the body or spread to other organs through a process called metastasis. Metastases are the leading cause of cancer death [1].
Cancer is influenced by several factors including the activation of proto-oncogenes through chromosomal translocations such as the presence of the Philadelphia chromosome, which is the result of the translocation of the BCR-ABL genes present in most patients with chronic myeloid leukemia (CML) [2]. Additionally, mutations can inactivate genes that give rise to cancer suppressors [3], for example, the mutation and consequent inactivation of p53, which leads to an accumulation of mutant protein, making it a hallmark of cancer [4]. Several miRNAs have been functionally classified as oncomiRs or tumor repressors because they act as transcriptional regulatory factors, depending on the target they regulate [5].
2. MiR-7 in Cancer
After the analysis carried out by the Lagos–Quintana group in 2001 where miRNAs 1 to 33 were studied, some of these were suggested to play an important role in the development of cancer, as well as act in a tissue-specific manner. miR-7 was mentioned for the first time in this publication [6] and was later reported to be encoded in three different loci: 9q21, 15q26, and 19q13, giving rise to three products, pri-miR-7-1, pri-miR-7-2, and pri-miR-7-3, which end in the same sequence as mature miR-7, as shown in Figure 1. Furthermore, it is known to be evolutionarily conserved [7].
Figure 1. miR-7 sequence. Representation of the microRNA-7 sequence obtained from miRDB (http://mirdb.org/cgi-bin/mature_mir.cgi?name=hsa-miR-7-5p accessed on 1 June 2022). The seed region is highlighted in red.
miR-7 has a high expression in healthy brain, spleen, and pancreas tissue [8]. Several reports have described this miRNA’s mechanisms of regulation; one of them indicated that the transcription factor HOXD10 is capable of interacting with the promoter region of the putative miR-7 -1 gene to induce the expression of miR-7 [9]. Another transcription factor involved is FOXP3, which induces the expression of miR-7 [10]. Thus, it has also been reported that c-Myc is positively regulates the expression of miR-7 by binding its promoter region [11]. Other proteins involved in pathways such as splicing factors were shown to have regulatory activity toward miR-7, such as the SD2/ASf factor, which interacts with the pri-miR-7 transcript to promote cleavage by Drosha [12]. Mechanisms involved in the immune response were also related to the regulation of miR-7 expression; such is the case of TLR9, the signaling of which increases the expression of HuR, increasing the expression of miR-7 in turn [13]. A regulation of TLR9 by miR-7 was observed in lung cancer, affecting the regulation of the PIK3R3/AKT pathway [14].
On the other hand, a study carried out on various types of cancer indicated that the promotion of the expression of miR-7 together with the reduction in the expression of miR-21, both by viral vectors, resulted in an antitumor effect that encompasses invasion, migration, cell proliferation, and increased survival [15]. In this way, the role of miR-7 in the development of different types of cancer was documented, as shown in Table 1.
Table 1. miR-7 targets and functions of target genes in cancer. Taken and modified from “microRNA-7: A tumor suppressor miRNA with therapeutic potential” [7].
| Target | Function | Type of Cancer | References |
|---|---|---|---|
| EGFR | Promotes cell survival, proliferation, tumorigenesis, resistance to therapeutic targets and radiotherapy | Head and neck, Glioblastoma multiforme, cervical, lung, breast, and prostate | [2][3][4][5][16] |
| RAF1 | Promotes cell survival | Lung, breast, and HNC | [3][5][17] |
| PAK1 | Promotes cell survival, proliferation, cell motility, invasion, growth, and tumorigenesis | Breast, squamous cell carcinoma of the tongue, Schwannoma | [16][18][19] |
| IRS-1 | Promotes proliferation | Glioblastoma multiforme, breast and cervical | [6][18] |
| IRS-2 | Promotes cell survival, proliferation, motility, and invasion | Glioblastoma multiforme, lung, breast, prostate, squamous cell carcinoma of the tongue, and Schwannoma melanoma | [3][6][18][19][20] |
| ACK1 | Promotes cell proliferation, and tumorigenesis | Schwannoma | [16] |
| PI3KCD | Promotes cell survival, proliferation, tumorigenesis, and metastasis | Hepatocellular carcinoma | [21] |
| mTOR | Promotes cell survival, proliferation, tumorigenesis, and metastasis | Hepatocellular carcinoma | [21] |
| P70S6K | Promotes cell survival, proliferation, tumorigenesis, and metastasis | Hepatocellular carcinoma | [21] |
| BCL-2 | Promotes resistance to apoptosis, proliferation, and tumorigenesis | Lung | [22] |
| XIAP | Promotes cell survival and proliferation | Cervical | [23] |
| YY1 | Promotes cell survival, proliferation, and tumorigenesis | Colorectal | [24] |
| CCNE1 | Promotes cell survival | Hepatocellular carcinoma | [24][25] |
| PA28γ gamma | Promotes cell survival, proliferation, and tumorigenesis | Lung | [26] |
| FAK | Promotes cell proliferation, cell survival, tumorigenesis, and cell mobility and regulates EMT | Glioblastoma multiforme and breast | [27][28] |
| KLF4 | Promotes metastasis and self-renewal of stem-type cancer cells | Breast | [29] |
| IGF1R | Promotes cell survival, proliferation, migration, invasion, and metastasis | Squamous cell carcinoma of the tongue, gastric | [19][30] |
| MRP1 | Promotes resistance to chemotherapy | Breast | [31] |
| ERF | Represses checkpoints in the cell cycle | Lung | [32] |
2.1. MiR-7 in Lung Cancer
Lung cancer is mainly composed of small cell and non-small cell cancer based on its microscopic appearance, with non-small cell cancer (NSCLC) being the most frequent. It is worth mentioning that it is the leading cause of cancer deaths in the United States [33]. In NSCLC, miR-7 was related to the ability to inhibit tumor growth. In a murine model, it was shown that tumor growth and metastasis of lung cancer cells were significantly reduced after inducing the expression of miR-7, which caused a reduction in the signaling of the Akt and ERK in pathways. Furthermore, the downregulation of NDUFA4, a new target of miR-7, was shown to contribute to the same effects of miR-7 expression generated by the TTF-1 promoter [34]. The same effect was demonstrated using drugs that had previously been effective in treating some types of cancer, excluding lung cancer—specifically NSCLC—in results published by Zeng’s group. They evaluated lung cancer cell lines (A549, NCL-H460) and A549 transfected with a mimic or an inhibitor of miR-7 and demonstrated that the compound Breviscapine (BVP), which is a crude extract of erigeron (Erigeron breviscapuos) and contains a large number of flavonoids, significantly reduced the growth of lung cancer cell lines and induced apoptosis. In addition, this entry proved that cells treated with the drug showed elevated levels of Bax and miR-7. This suggested that the elevation in miR-7 expression promotes the BVP sensitivity of NSCLC cells by suppressing proliferation and promoting apoptosis [35]. Another drug that was shown to increase miR-7 is docetaxel, an antineoplastic drug defined as an antimitotic agent that inhibits the formation of microtubules. In NSCLC cell lines, docetaxel increases the expression of miR-7, and, in turn, this overexpression suppresses proliferation in vitro [36]. The expression of miR-7 also induces inhibition of cancer cell growth, which may be associated with the decrease in the expression of the protein associated with tumor growth CGGBP1, one of the reported targets of miR-7 [37].
Some other targets specifically reported in non-small cell lung cancer are PAX6, a highly conserved transcription factor implicated in NSCLC; miR-7 induces the inhibition of PAX6 expression, thereby reducing the proliferation potential, migration, and invasion of NSCLC cells via ERK/MAPK [38]. In addition, miR-7 has been reported to inhibit FAK, negative regulation of which is also reflected in inhibiting the activation of the ERK/MAPK pathway, for which both PAX6 and FAK were proposed as new targets for NSCLC treatment [39]. Another reported target is p28α, the overexpression of which has the same effect in NSCLC cell lines as miR-7 downregulation [40]. Thus, the reduced expression of miR-7 was not only found in cell lines, but also in NSCLC tissues [40], while in A549 cells, as well as in NSCLC tissue, miR-7 was shown to inhibit metastasis by NOVA2 targeting [41].
In elucidating the mechanism by which miR-7 acts in tumorigenesis, DNA sequencing has been utilized to determine whether there are mutation sites in the miR-7-2 promoter in lung cancer tissues, and changes such as GC at site -617 and an AG change at site -604 are associated with poor prognosis in cancer patients [42]. The abovementioned mechanisms are illustrated in Figure 2.
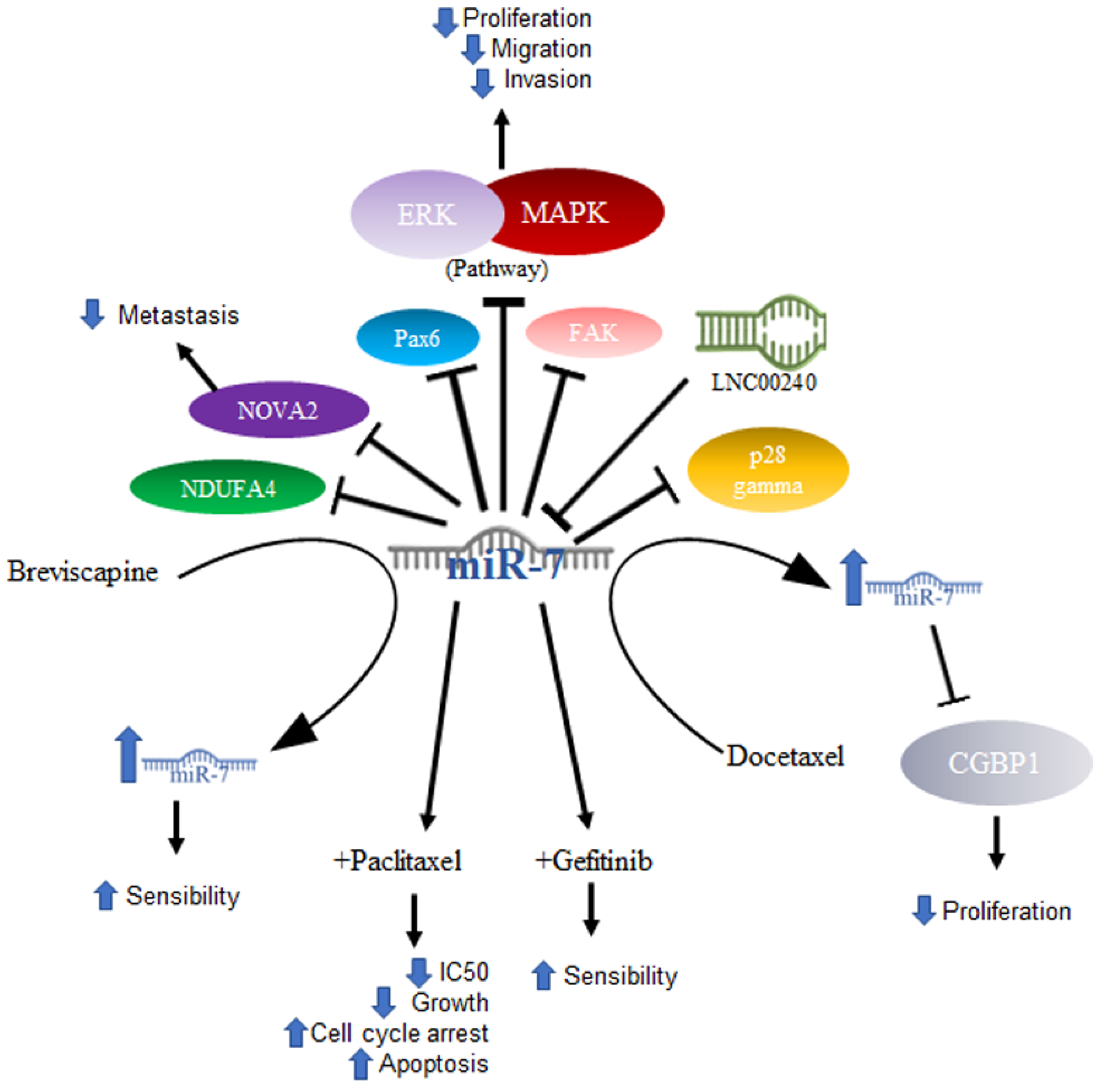
Figure 2. Role of miR-7 in lung cancer. Schematic representation of protein interactions, roles, and treatment with miR-7 and its biological consequences in lung cancer.
Another area under investigation is the role played by miR-7 in resistance to treatment. Studies conducted with paclitaxel (PTX), an antineoplastic drug that inhibits microtubule formation and is the drug of first choice in the treatment of NSCLC, demonstrated that overexpression of miR-7 in NSCLC cell lines increased sensitivity to paclitaxel, suppressed cell proliferation, and increased apoptosis. On the other hand, miR-7 inhibition decreased the anti-proliferative and pro-apoptotic effects of PTX, and miR-7 upregulation enhanced the PTX-sensitivity of NSCLC cells, probably owing to the downregulation of EGFR expression [43]. Data on gefitinib, another drug used in the treatment of NSCLC, which is a tyrosine kinase inhibitor targeting epidermal growth factor receptors or EGFRs, showed that gefitinib treatment coupled with miR-7 significantly decreased the IC50 of gefitinib, inhibited cell growth, increased G0/G1 cell cycle arrest, and increased apoptosis [44]. In a very interesting way and consistent with what was mentioned above, in a study carried out on lung cancer cell lines, it was shown that the lncRNA LNC00240 acts as a miR-7-5p sponge and, as a consequence, induces EGFR overexpression [45].
2.2. MiR-7 in Hepatocellular Cancer
Hepatocellular cancer (HCC) or liver cancer is more common in men than in women. Liver cancer can be classified as intrahepatic cholangiocarcinoma or bile duct cancer, angiosarcoma or hemangiosarcoma, or hepatoblastoma. HCC is usually successfully treated with chemotherapy and surgery; however, complications can occur and are more frequent when the cancer originates outside the liver [46].
In HCC, miR-7 is characterized as anti-oncogenic; however, the mechanism is not clear [47]. Studies have been published in which several mechanisms of action were proposed: one of them included the complex formed by nuclear factor 90 (NF90) and nuclear factor 45 (NF45), which acts as a negative regulator of miR-7 processing. The expression of NF90 and NF45 were found to be significantly elevated in primary tumors compared to adjacent non-tumor tissues. After the elimination of NF90 and analysis by miRNA microarray as well as quantitative RT-PCR, results included an elevated expression of mature miR-7 and the decreased expression of pri-miR-7-1 [48]. On the other hand, it has been reported that elements such as voltage-dependent anion channel 1 (VDAC1), which plays an important role in carcinogenesis, play prognostic roles in patients with hepatocellular cancer, and miR-7 was suggested to suppress VDAC1 expression [49]. Another factor related to a poor prognosis is maspin, the expression of which is reduced in patients with hepatitis B virus and correlates with a poor prognosis. Maspin is involved in tumor suppression by promoting cell adhesion and apoptosis, particularly in patients with hepatocellular carcinoma (HCC). Maspin is negatively regulated by hepatitis B virus protein x (HBx). High expressions of miR-7, 107, and 21 were related to the negative regulation of maspin, which was suggested to confer aggressiveness and chemoresistance mediated by HBx in HCC [50].
The involvement of miR-7 was also found in the cell cycle, since cyclin E1 (CCNE1) was identified as an important mediator of the G1/S transition, and reports indicated that this is a direct target of microRNA-7. The overexpression of miR-7 has the effect of silencing CCNE1, thus promoting the tumor suppressor effects attributed to the expression of the CCNE1 oncogene [51]. Finally, it was also reported that miR-7 is capable of negatively regulating SPC24 and, as a consequence, acting as a tumor inhibitor since it suppresses proliferation and migration and promotes apoptosis [52]. The abovementioned mechanisms are illustrated in Figure 3.
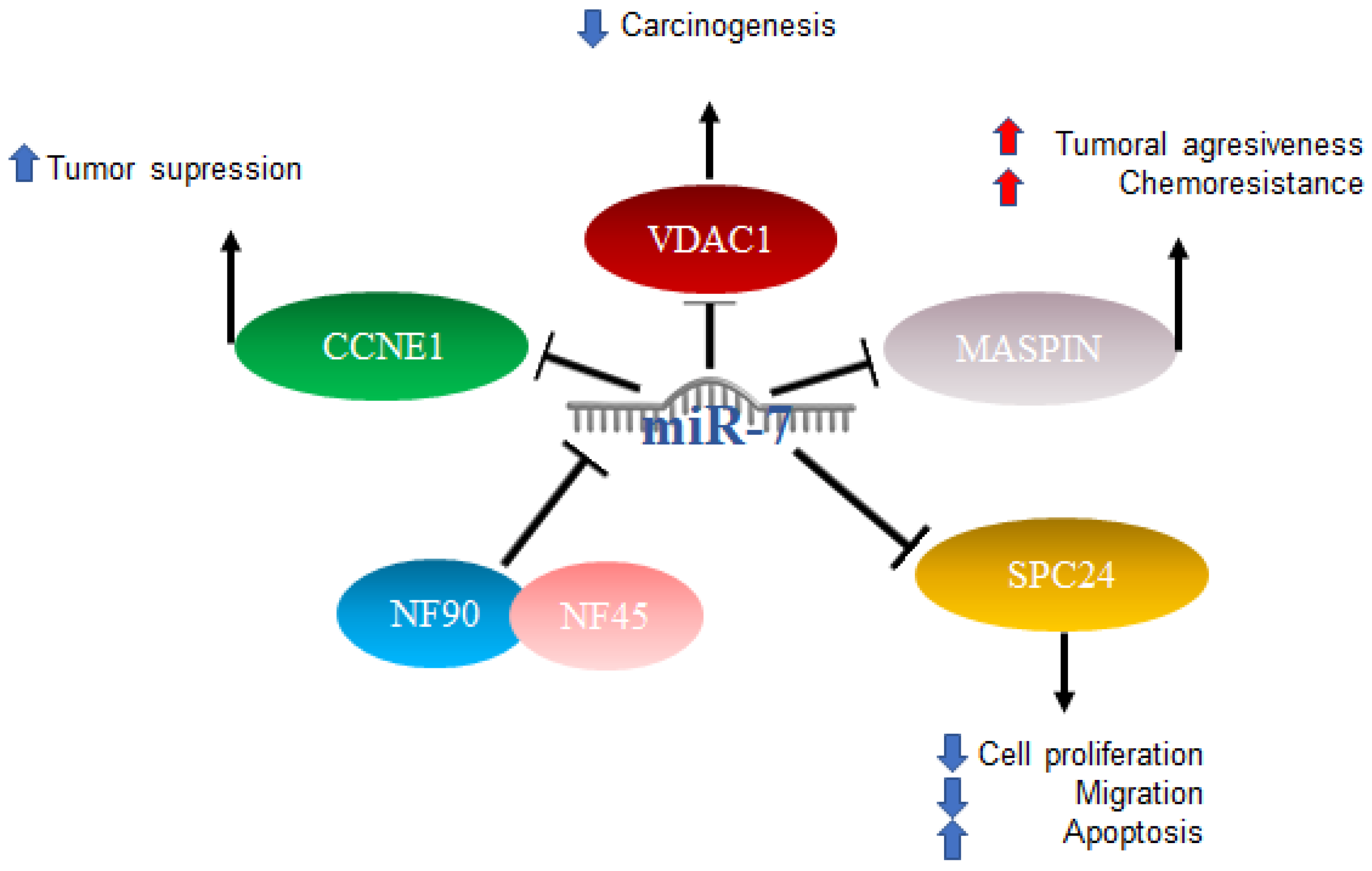
Figure 3. Role of miR-7 in hepatocellular cancer. Schematic representation of protein interactions and roles of miR-7 and its biological consequences in hepatocellular cancer.
2.3. MiR-7 in Breast Cancer
Breast cancer manifests itself as abnormal cell growth forming a tumor that can be felt as a lump. The tumor is considered malignant when it is capable of invading surrounding tissues or if it is capable of generating metastases. It is considered almost exclusively a disease of women; however, it can also occur in men, and early detection generally leads to a good prognosis [53].
Various roles for miR-7 have also been reported in breast cancer. Particularly in triple-negative patients, there is usually treatment failure. One of the main characteristics is the elevation in the expression of IL-6, and treatment with antibodies against IL-6 inhibits the migration induced by lapatinib, which is a drug that promotes metastasis in triple-negative breast cancer cells. RAF-1/MAPK, JNK‘s, p38 MAPK and AP-1 activator protein signaling mechanisms are activated in response to lapatinib treatment to induce IL-6 expression. It was reported that miR-7 binds to the 3′UTR region of RAF-1, inhibiting its activity, which leads to the negative regulation of this pathway by miR-7, acting as an antitumor mechanism [54].
miR-7 is also involved in the oncogenic isoform of the HER2 gene, known as HER2Δ16, which is present in 50% of HER2+ breast cancer patients. Reports have indicated that this isoform leads to metastasis and resistance to chemotherapy. It was recently shown that HER2Δ16 alters the expression of some miRNAs. Using an array comparison of miRNA expression profiles of the MCF/7 and MCF7/HER2Δ16 cell lines, 16 miRNAs were found to be significantly altered by two or more folds. miR-7 showed a greater increase, at 4.8-fold in the MCF7/HER2Δ16 lines. The restoration of miR-7 expression promotes G1 cell cycle arrest and reduces colony formation and cell migration to levels similar to MCF7, while the suppression of miR-7 in MCF7 results in promotion of colony formation, but not in cell migration. The mechanism involved includes EGFR and Src Kinase, which represent targets for therapeutic intervention and refraction and metastasis of breast cancer with HER2Δ16 [55].
Another signaling pathway in which miR-7 participates is WISP2 (WNT1-inducible-signaling pathway protein 2); silencing WISP2 signaling in the MCF7 cells of human breast adenocarcinoma impairs cytotoxic T-lymphocyte (CTL)-mediated cell death through a mechanism that involves the induction of Kruppel-like factor-4 (KLF-4) and miR-7 decrease. Using a WISP2 inhibitor results in a significant reversal of the epithelial-to-mesenchymal transition. KLF4 expression was found to be related to the inhibition of miR-7, which is responsible for CTL-mediated lysis. WISP2 has a role in tumor susceptibility through EMT via the TGF-β signaling pathway, KLF4 expression, and miR-7 inhibition, thus proposing WISP2 as an activator of CTL-induced death and suggesting that the loss of its function promotes immune surveillance evasion and tumor promotion [56]. On the other hand, in stem-type breast cancer cell lines with metastatic capacity to the brain, a profile of miRNAs was made that showed a reduced expression of miR-7, which promotes the expression of KLF4 factor in induced pluripotent stem cells. It was reported that miR-7 expression can decrease KLF4 expression and suppress breast cancer cell metastasis [57].
Another report indicated that in breast cancer cell lines and through qRT-PCR, a high expression of miR-7 is directly related to a better response to chemotherapy with paclitaxel/carboplatin through the suppression of MRP1 and BCL2 [58]. The mechanisms mentioned above are illustrated in Figure 4.
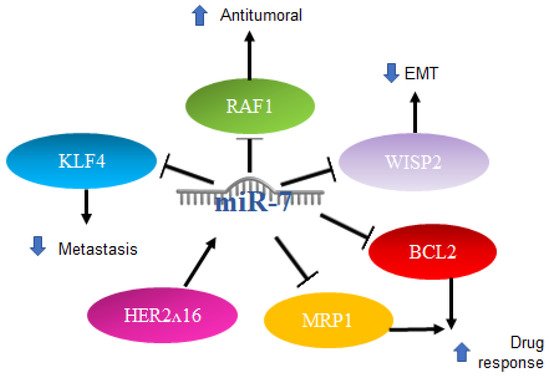
Figure 4. Role of miR-7 in breast cancer. Schematic representation of protein interactions and roles of miR-7 and its biological consequences in breast cancer.
2.4. Role of MiR-7 in Gliomas
Gliomas are among the ten most common causes of death related to cancer. Although there is no test to detect it early, timely diagnosis and treatment improve the outcome. Recent progress has been made in the scientific understanding and in methods of diagnosis and treatment [59]. Alterations in energy metabolism were observed in gliomas involving Akt signaling in aerobic glycolysis programs; however, the mechanisms regulating aerobic glycolysis and Akt activity are not known. One of the proposed mechanisms includes the regulation of IGF-1R, which is part of the signaling pathway upstream of Akt. The increase in the expression of miR-7 was reported to inhibit colony formation and the glucose metabolic capacity of glioma cells, an effect similar to that produced by the “knockdown” of IGF-R, which is consistent with data indicating that miR-7 is capable of binding to the 3′UTR region of IGF-R, causing its decrease. Expression data of miRNAs downloaded from the Genomic Atlas of Cancer database were analyzed [60]. Thus, the expression profiles of 20 mature miRNAs were also studied in patient biopsies with the finding that miR-7 is less expressed in tumors compared to healthy control samples [61]. In addition to the Akt pathway, miR-7 is involved in the regulation of the RAF/ampk (MEK) and ERK pathways, two of which are initiated by EGFR, which in glioblastoma is reported to be a regulator of cell proliferation, survival, migration, and invasion. An increase in the expression of miR-7 decreases the expression of PI3K, phosphorylation of AKT, RAF-1, and phosphorylated MEK1/2, and causes a slight reduction in the expression of EGFR. An induced expression of PI3K or RAF-1 reverses the effects of miR-7. Therefore, miR-7 was proposed as a potential tumor suppressor because it is capable of regulating several related oncogenes in the EGFR pathway [62]. Additionally, the overexpression of miR-7-5p was found capable of inhibiting SATB1 (SATB homeobox 1) and reversing the effects of promoting cell migration and invasion in glioblastoma cells [63]. Another target of miR-7-5p in glioblastoma is YY1 (Ying Yang 1), the silencing of which has direct effects on the reduction in the chemoresistance of cells to temozolomide [64].
Furthermore, the first trials using miRNA as part of a therapy were performed with promising results; the administration of miR-7 encapsulated in cationic liposomes in xenografts arrested growth, and metastatic nodules decreased the effectiveness in a sequence-specific manner. miR-7 has been applied in the treatment of glioma, and the results have shown it to be a promising potential anti-tumor and anti-metastatic treatment for human glioma [65]. All the mechanisms mentioned above are illustrated in Figure 5.
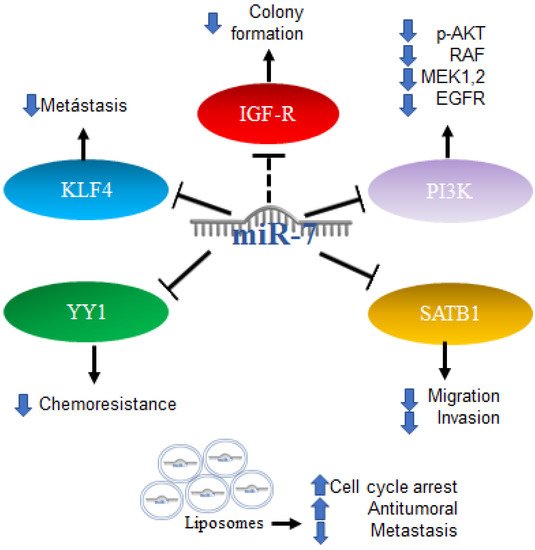
Figure 5. Role of miR-7 in gliomas. Schematic representation of protein interactions, roles and treatment with miR-7 and its biological consequences in gliomas.
2.5. Role of MiR-7 in Colorectal Cancer
Colorectal cancer (CRC) can occur in the colon or rectum, hence the name; most cases of this type of cancer begin with a growth called a polyp in the inner lining of the colon. Adenocarcinomas represent about 95% of colorectal cancers [66]. Colorectal cancer studies conducted on 196 malignant and 14 normal tissues, analyzing the expression of miR-7 levels with real-time PCR, found that miR-7 was significantly elevated in colorectal cancer compared to normal colorectal mucosa. From this entry, it was determined that a high expression of miR-7 is significantly associated with poor survival in patients with CRC (p = 0.01) [67]. In addition, it was determined by in vitro assays that EGFR and RAF-1 are targets of miR-7 [68]. Additionally, it was reported in colorectal cancer that the 3′UTR region of YY1 had binding sites for miR-7, which was confirmed by reporter plasmid assays, and that miR-7 was downregulated in colorectal cancer cell lines and in the ectopic expression results on the suppression of cancer cell proliferation, apoptosis induction and cell cycle arrest. In parallel, the “knockdown” of YY1 suppresses cell proliferation and induces apoptosis, indicating the inverse functions of miR-7 and YY1 in CRC cell lines, where YY1 promotes CRC proliferation and miR-7 inhibits it by inhibiting YY1 expression. Based on this, miR-7 was proposed as a tumor suppressor by suppressing YY1 in its oncogenic role. YY1 can promote cell growth by inhibiting p53 and promoting the Wnt signaling pathway [69]. Therefore, the precursor of miR-7 was proposed as a possible therapy. In contrast, in another study carried out on CRC tissues and cells, it was determined that a low expression of miR-7-5p in CRC tissue predicts a low 5-year survival, while in cells, it was determined that the overexpression of this microRNA is directly related to the inhibition of cell proliferation and migration. This represents a controversial finding on the expression of miR-7 in CRC, which should be addressed and based on the targets of miR-7 in this context. Additionally, it was reported that KLF4, a target of miR-7, acts as an oncogene in colorectal cancer, so the inhibition of KLF4 expression by miR-7 is a mechanism and a potential molecular target for the treatment of colorectal cancer [70]. TRIP6 is another target of miR-7 that was related to the regulation of metastasis and inhibition of cell proliferation; however, in colorectal cancer cells, it was reported that miR-7 is decreased as a consequence of hypermethylation of TRIP6, while the restoration of miR-7 expression has a direct impact on the inhibition of cell proliferation and migration [71]. Interestingly, it was reported that, in biopsies of patients diagnosed with colorectal cancer, the expression of miR-7-5p together with miR-10a-5p can serve as a marker of poor prognosis [72]. The mechanisms mentioned above are illustrated in Figure 6.
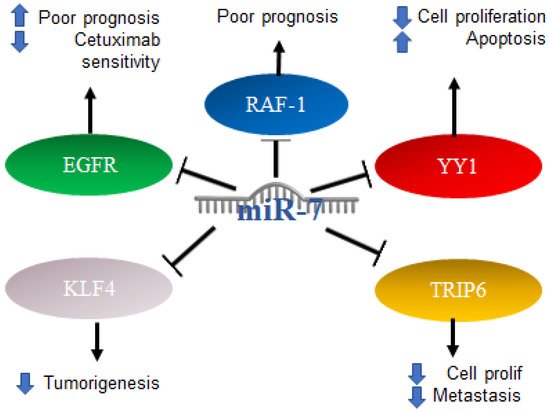
Figure 6. Role of miR-7 in colorectal cancer. Schematic representation of protein interactions with miR-7 and its biological consequences in colorectal cancer.
2.6. Role of MiR-7 in Prostate Cancer
The prostate is a gland that only men have, and the abnormal growth of the cells generates prostate cancer, which alters the gland’s growth. The prostate is normally compared to a walnut in regard to size. Prostate cancer is more common in men of 40 years or older [73].
miR-7 has been described as a tumor suppressor capable of annulling the stemness of prostate cancer cells, inhibiting tumorigenesis by suppressing KLF4. The possible mechanism is the regulation of the KLF4/PI3K/Akt/p21 pathways [74]. Studies were conducted on PC3 and LnCap prostate cancer cell lines, where it was found by means of an expression profile that miR-7 is increased in PC3 (11.3 times increased, p = 0.012). However, in patients with a high expression of miR-7 in peripheral blood, a significantly decreased survival rate was observed. Therefore, miR-7 is proposed as a potential biomarker in the prognosis of prostate cancer [75].
References
- OMS. “Cáncer,” Fact Sheets, 2017. Available online: http://www.who.int/mediacentre/factsheets/fs297/es/ (accessed on 6 December 2017).
- Reckel, S.; Hantschel, O. Bcr-Abl: One kinase, two isoforms, two diseases. Oncotarget 2017, 8, 78257–78258.
- Koeffler, H.P.; McCormick, F.; Denny, C. Molecular mechanisms of cancer. West. J. Med. 1991, 155, 505–514.
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474.
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587.
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858.
- Kalinowski, F.; Brown, R.A.; Ganda, C.; Giles, K.M.; Epis, M.R.; Horsham, J.; Leedman, P.J. microRNA-7: A tumor suppressor miRNA with therapeutic potential. Int. J. Biochem. Cell Biol. 2014, 54, 312–317.
- Correa-Medina, M.; Bravo-Egana, V.; Rosero, S.; Ricordi, C.; Edlund, H.; Diez, J.; Pastori, R.L. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr. Patterns 2009, 9, 193–199.
- Reddy, S.D.N.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-7, a Homeobox D10 Target, Inhibits p21-Activated Kinase 1 and Regulates Its Functions. Cancer Res. 2008, 68, 8195–8200.
- McInnes, N.; Sadlon, T.J.; Brown, C.Y.; Pederson, S.; Beyer, M.; Schultze, J.L.; McColl, S.; Goodall, G.J.; Barry, S.C. FOXP3 and FOXP3-regulated microRNAs suppress SATB1 in breast cancer cells. Oncogene 2011, 31, 1045–1054.
- Chou, Y.-T.; Lin, H.-H.; Lien, Y.-C.; Wang, Y.-H.; Hong, C.-F.; Kao, Y.-R.; Lin, S.-C.; Chang, Y.-C.; Lin, S.-Y.; Chen, S.-J.; et al. EGFR Promotes Lung Tumorigenesis by Activating miR-7 through a Ras/ERK/Myc Pathway That Targets the Ets2 Transcriptional Repressor ERF. Cancer Res. 2010, 70, 8822–8831.
- Wu, H.; Sun, S.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.; Zhu, J. A Splicing-Independent Function of SF2/ASF in MicroRNA Processing. Mol. Cell 2010, 38, 67–77.
- Li, Y.-J.; Wang, C.-H.; Zhou, Y.; Liao, Z.-Y.; Zhu, S.-F.; Hu, Y.; Chen, C.; Luo, J.-M.; Wen, Z.-K.; Xu, L. TLR9 signaling repressed tumor suppressor miR-7 expression through up-regulation of HuR in human lung cancer cells. Cancer Cell Int. 2013, 13, 90.
- Xu, L.; Wen, Z.; Zhou, Y.; Liu, Z.; Li, Q.; Fei, G.; Luo, J.; Ren, T. MicroRNA-7–regulated TLR9 signaling–enhanced growth and metastatic potential of human lung cancer cells by altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt pathway. Mol. Biol. Cell 2013, 24, 42–55.
- Bhere, D.; Arghiani, N.; Lechtich, E.R.; Yao, Y.; Alsaab, S.; Bei, F.; Matin, M.M.; Shah, K. Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy. Sci. Rep. 2020, 10, 1–10.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862.
- Ambros, V. microRNAs: Tiny Regulators with Great Potential. Cell 2001, 107, 823–826.
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864.
- MLagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739.
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728.
- Dostie, J.; Mourelatos, Z.; Yang, M.; Sharma, A.; Dreyfuss, G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003, 9, 180–186.
- Ambros, V. MicroRNA Pathways in Flies and Worms: Growth, Death, Fat, Stress, and Timing. Cell 2003, 113, 673–676.
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila melanogaster Development. Dev. Cell 2003, 5, 337–350.
- Lim, L.P.; Glasner, M.E.; Yekta, S.; Burge, C.B.; Bartel, D.P. Vertebrate MicroRNA Genes. Science 2003, 299, 1540.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Denli, A.M.; Tops, B.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235.
- Basyuk, E.; Suavet, F.; Doglio, A.; Bordonné, R.; Bertrand, E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003, 31, 6593–6597.
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296.
- Hutvágner, G.; Mclachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838.
- American-Cancer-Society, “American Cancer Society, Cancer facts and figures 2015”. 2015. Available online: www.cancer.org (accessed on 1 June 2022).
- Lei, L.; Chen, C.; Zhao, J.; Wang, H.; Guo, M.; Zhou, Y.; Luo, J.; Zhang, J.; Xu, L. Targeted Expression of miR-7 Operated by TTF-1 Promoter Inhibited the Growth of Human Lung Cancer through the NDUFA4 Pathway. Mol. Ther.-Nucleic Acids 2017, 6, 183–197.
- Zeng, J.; Cai, S. Breviscapine suppresses the growth of non-small cell lung cancer by enhancing microRNA-7 expression. J. Biosci. 2017, 42, 121–129.
- He, X.; Li, C.; Wu, X.; Yang, G. Docetaxel inhibits the proliferation of non-small-cell lung cancer cells via upregulation of microRNA-7 expression. Int. J. Clin. Exp. Pathol. 2015, 8, 9072–9080.
- Hu, Y.; Liao, Z.; Chen, C.; Qin, N.; Zheng, J.; Tian, D.; Li, Y.; Zhu, S.; Luo, J.; Xu, L. Over-expressed microRNA-7 inhibits the growth of human lung cancer cells via suppressing CGGBP1 expression. Chin. J. Cell. Mol. Immunol. 2014, 30, 125–130.
- Luo, J.; Li, H.; Zhang, C. MicroRNA-7 inhibits the malignant phenotypes of non-small cell lung cancer in vitro by targeting Pax6. Mol. Med. Rep. 2015, 12, 5443–5448.
- Cao, Q.; Mao, Z.-D.; Shi, Y.-J.; Chen, Y.; Sun, Y.; Zhang, Q.; Song, L.; Peng, L.-P. MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget 2016, 7, 77468–77481.
- Xiong, S.; Zheng, Y.; Jiang, P.; Liu, R.; Liu, X.; Qian, J.; Gu, J.; Chang, L.; Ge, D.; Chu, Y. PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br. J. Cancer 2013, 110, 353–362.
- Xiao, H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell. Mol. Biol. Lett. 2019, 24, 1–13.
- Zhao, J.; Wang, K.; Liao, Z.; Li, Y.; Yang, H.; Chen, C.; Zhou, Y.; Tao, Y.; Guo, M.; Ren, T.; et al. Promoter mutation of tumor suppressor microRNA-7 is associated with poor prognosis of lung cancer. Mol. Clin. Oncol. 2015, 3, 1329–1336.
- Liu, R.; Liu, X.; Zheng, Y.; Gu, J.; Xiong, S.; Jiang, P.; Jiang, X.; Huang, E.; Yang, Y.; Ge, D.; et al. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncol. Lett. 2014, 8, 2193–2200.
- Zhao, J.-G.; Men, W.-F.; Tang, J. MicroRNA-7 enhances cytotoxicity induced by gefitinib in non-small cell lung cancer via inhibiting the EGFR and IGF1R signalling pathways. Współczesna Onkol. 2015, 3, 201–206.
- Ku, G.W.; Kang, Y.; Yu, S.-L.; Park, J.; Park, S.; Jeong, I.B.; Kang, M.W.; Son, J.W.; Kang, J. LncRNA LINC00240 suppresses invasion and migration in non-small cell lung cancer by sponging miR-7-5p. BMC Cancer 2021, 21, 44.
- American-Cancer-Society. Acerca del Cáncer de Hígado, 2016. Available online: https://www.cancer.org/es/cancer/cancer-de-higado/acerca/que-es-cancer-de-higado.html (accessed on 1 June 2022).
- Fang, Y.; Xue, J.-L.; Shen, Q.; Chen, J.; Tian, L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012, 55, 1852–1862.
- Higuchi, T.; Todaka, H.; Sugiyama, Y.; Ono, M.; Tamaki, N.; Hatano, E.; Takezaki, Y.; Hanazaki, K.; Miwa, T.; Lai, S.; et al. Suppression of MicroRNA-7 (miR-7) Biogenesis by Nuclear Factor 90-Nuclear Factor 45 Complex (NF90-NF45) Controls Cell Proliferation in Hepatocellular Carcinoma. J. Biol. Chem. 2016, 291, 21074–21084.
- Wang, F.; Qiang, Y.; Zhu, L.; Jiang, Y.; Wang, Y.; Shao, X.; Yin, L.; Chen, J.; Chen, Z. MicroRNA-7 downregulates the oncogene VDAC1 to influence hepatocellular carcinoma proliferation and metastasis. Tumor Biol. 2016, 37, 10235–10246.
- Chen, W.-S.; Yen, C.-J.; Chen, Y.-J.; Chen, J.-Y.; Wang, L.-Y.; Chiu, S.-J.; Shih, W.-L.; Ho, C.-Y.; Wei, T.-T.; Pan, H.-L.; et al. miRNA-7/21/107 contribute to HBx-induced hepatocellular carcinoma progression through suppression of maspin. Oncotarget 2015, 6, 25962–25974.
- Zhang, X.; Hu, S.; Zhang, X.; Wang, L.; Zhang, X.; Yan, B.; Zhao, J.; Yang, A.; Zhang, R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2013, 443, 1078–1084.
- Wang, Y.; Yang, H.; Zhang, G.; Luo, C.; Zhang, S.; Luo, R.; Deng, B. hsa-miR-7-5p suppresses proliferation, migration and promotes apoptosis in hepatocellular carcinoma cell lines by inhibiting SPC24 expression. Biochem. Biophys. Res. Commun. 2021, 561, 80–87.
- American-Cancer-Society. Que es el Cancer de Seno, 2016. Available online: https://www.cancer.org/es/cancer/cancer-de-seno/acerca/que-es-el-cancer-de-seno.html (accessed on 1 June 2022).
- Hsiao, Y.-C.; Yeh, M.-H.; Chen, Y.-J.; Liu, J.-F.; Tang, C.-H.; Huang, W.-C. Lapatinib increases motility of triple-negative breast cancer cells by decreasing miRNA-7 and inducing Raf-1/MAPK-dependent interleukin-6. Oncotarget 2015, 6, 37965–37978.
- Huynh, F.C.; Jones, F.E. MicroRNA-7 Inhibits Multiple Oncogenic Pathways to Suppress HER2Δ16 Mediated Breast Tumorigenesis and Reverse Trastuzumab Resistance. PLoS ONE 2014, 9, e114419.
- Akalay, I.; Tan, T.Z.; Kumar, P.; Janji, B.; Mami-Chouaib, F.; Charpy, C.; Vielh, P.; Larsen, A.K.; Thiery, J.P.; Sabbah, M.; et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene 2014, 34, 2261–2271.
- Okuda, H.; Xing, F.; Pandey, P.R.; Sharma, S.; Watabe, M.; Pai, S.K.; Mo, Y.-Y.; Iiizumi-Gairani, M.; Hirota, S.; Liu, Y.; et al. miR-7 Suppresses Brain Metastasis of Breast Cancer Stem-Like Cells By Modulating KLF4. Cancer Res. 2013, 73, 1434–1444.
- Hong, T.; Ding, J.; Li, W. miR-7 Reverses Breast Cancer Resistance to Chemotherapy by Targeting MRP1 and BCL2. OncoTargets Ther. 2019, 12, 11097–11105.
- Schneider, T.; Mawrin, C.; Scherlach, C.; Skalej, M.; Firsching, R. Gliomas in Adults. Dtsch. Arztebl. Int. 2010, 107, 799–808.
- Wang, B.; Sun, F.; Dong, N.; Sun, Z.; Diao, Y.; Zheng, C.; Sun, J.; Yang, Y.; Jiang, D. MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn. Pathol. 2014, 9, 211.
- Koshkin, F.A.; Chistyakov, D.A.; Nikitin, A.G.; Konovalov, A.N.; Potapov, A.; Usachyov, D.Y.; Pitskhelauri, D.I.; Kobyakov, G.L.; Shishkina, L.V.; Chekhonin, V.P. Profile of MicroRNA Expression in Brain Tumors of Different Malignancy. Bull. Exp. Biol. Med. 2014, 157, 794–797.
- Liu, Z.; Jiang, Z.; Huang, J.; Huang, S.; Li, Y.; Yu, S.; Yu, S.; Liu, X. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 2014, 44, 1571–1580.
- Yin, C.; Kong, W.; Jiang, J.; Xu, H.; Zhao, W. miR-7-5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Oncol. Lett. 2018, 17, 1819–1825.
- Jia, B.; Liu, W.; Gu, J.; Wang, J.; Lv, W.; Zhang, W.; Hao, Q.; Pang, Z.; Mu, N.; Zhang, W.; et al. MiR-7-5p suppresses stemness and enhances temozolomide sensitivity of drug-resistant glioblastoma cells by targeting Yin Yang 1. Exp. Cell Res. 2018, 375, 73–81.
- Wang, W.; Dai, L.X.; Zhang, S.; Yang, Y.; Yan, N.; Fan, P.; Dai, L.; Tian, H.W.; Cheng, L.; Zhang, X.M.; et al. Regulation of Epidermal Growth Factor Receptor Signaling by plasmid-based MicroRNA-7 inhibits human malignant gliomas growth and metastasis in vivo. Neoplasma 2013, 60, 274–283.
- American-Cancer-Society. Acerca del Cáncer de Colon y Recto, 2016. Available online: https://www.cancer.org/es/cancer/cancer-de-colon-o-recto/acerca/que-es-cancer-de-colon-o-recto.html (accessed on 1 June 2022).
- Nagano, Y.; Toiyama, Y.; Okugawa, Y.; Imaoka, H.; Fujikawa, H.; Yasuda, H.; Yoshiyama, S.; Hiro, J.; Kobayashi, M.; Ohi, M.; et al. MicroRNA-7 Is Associated with Malignant Potential and Poor Prognosis in Human Colorectal Cancer. Anticancer Res. 2016, 36, 6521–6526.
- Suto, T.; Yokobori, T.; Yajima, R.; Morita, H.; Fujii, T.; Yamaguchi, S.; Altan, B.; Tsutsumi, S.; Asao, T.; Kuwano, H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2014, 36, 338–345.
- Zhang, N.; Li, X.; Wu, C.W.; Dong, Y.; Cai, M.; Mok, M.T.S.; Wang, H.; Chen, J.; Ng, S.S.M.; Chen, M.; et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 2012, 32, 5078–5088.
- Dong, M.; Xie, Y.; Xu, Y. miR-7-5p regulates the proliferation and migration of colorectal cancer cells by negatively regulating the expression of Krüppel-like factor 4. Oncol. Lett. 2019, 17, 3241–3246.
- Ling, Y.; Cao, C.; Li, S.; Qiu, M.; Shen, G.; Chen, Z.; Yao, F.; Chen, W. TRIP6, as a target of miR-7, regulates the proliferation and metastasis of colorectal cancer cells. Biochem. Biophys. Res. Commun. 2019, 514, 231–238.
- Romero-Lorca, A.; Novillo, A.; Gaibar, M.; Gilsanz, M.F.; Galán, M.; Beltrán, L.; Antón, B.; Malón, D.; Moreno, A.; Fernández-Santander, A. miR-7, miR-10a and miR-143 Expression May Predict Response to Bevacizumab Plus Chemotherapy in Patients with Metastatic Colorectal Cancer. Pharm. Pers. Med. 2021, 14, 1263–1273.
- American-Cancer-Society. Acerca del Cáncer de Próstata, 2016. Available online: https://www.cancer.org/es/cancer/cancer-de-prostata/acerca/que-es-cancer-de-prostata.html (accessed on 1 June 2022).
- Chang, Y.-L.; Zhou, P.-J.; Wei, L.; Li, W.; Ji, Z.; Fang, Y.-X.; Gao, W.-Q. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget 2015, 6, 24017–24031.
- Santos, J.I.; Teixeira, A.L.; Dias, F.; Maurício, J.; Lobo, F.; Morais, A.; Medeiros, R. Influence of peripheral whole-blood microRNA-7 and microRNA-221 high expression levels on the acquisition of castration-resistant prostate cancer: Evidences from in vitro and in vivo studies. Tumor Biol. 2014, 35, 7105–7113.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Entry Collection:
Chemical Bond
Revisions:
2 times
(View History)
Update Date:
16 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




