
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Aiping Song | -- | 2711 | 2022-08-15 14:27:38 | | | |
| 2 | Beatrix Zheng | Meta information modification | 2711 | 2022-08-16 05:03:38 | | | | |
| 3 | Beatrix Zheng | + 2 word(s) | 2713 | 2022-08-16 05:09:46 | | |
Video Upload Options
Transcription factors, also known as trans-acting factors, balance development and stress responses in plants. Branching plays an important role in plant morphogenesis and is closely related to plant biomass and crop yield. The apical meristem produced during plant embryonic development repeatedly produces the body of the plant, and the final aerial structure is regulated by the branching mode generated by axillary meristem (AM) activities. These branching patterns are regulated by two processes: AM formation and axillary bud growth. In recent years, transcription factors involved in regulating these processes have been identified. In addition, these transcription factors play an important role in various plant hormone pathways and photoresponses regulating plant branching.
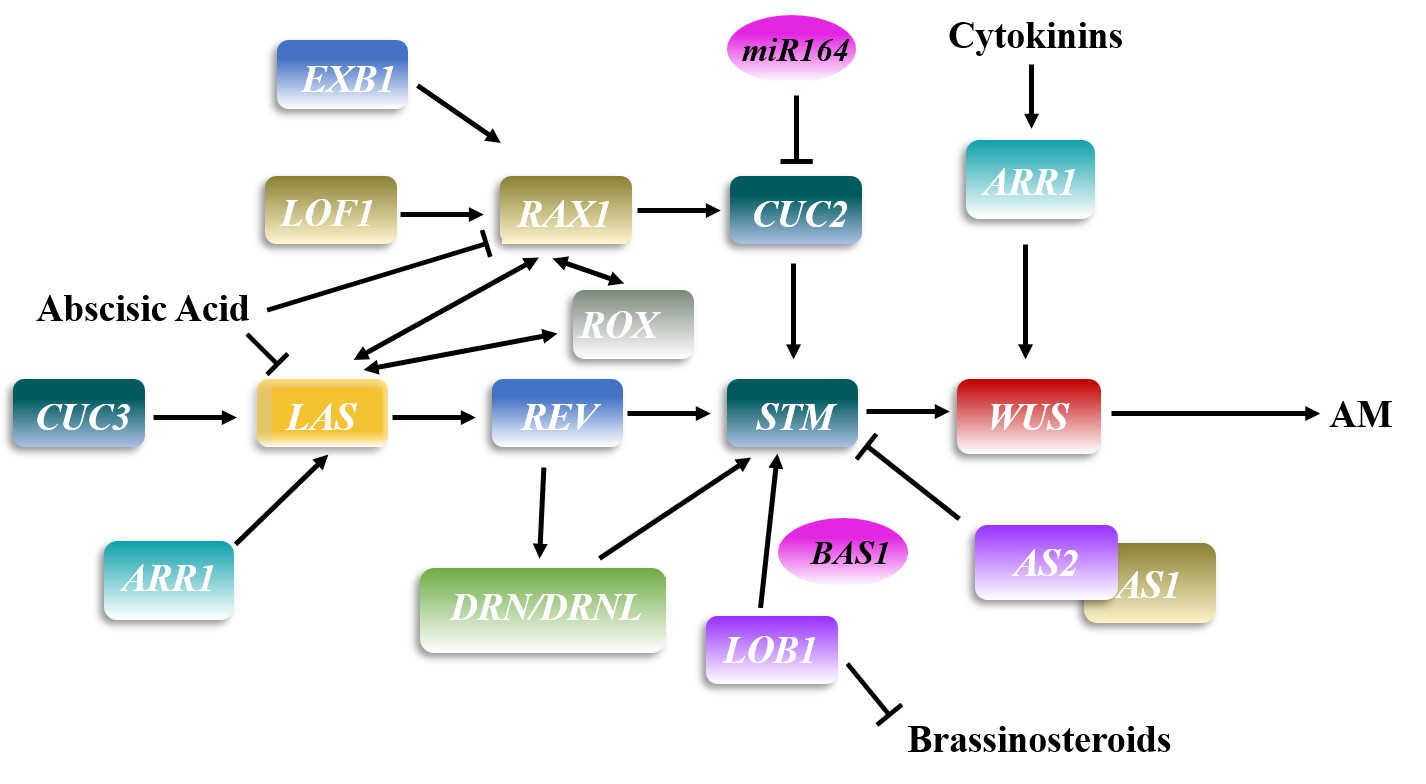
1. Axillary Meristem Formation
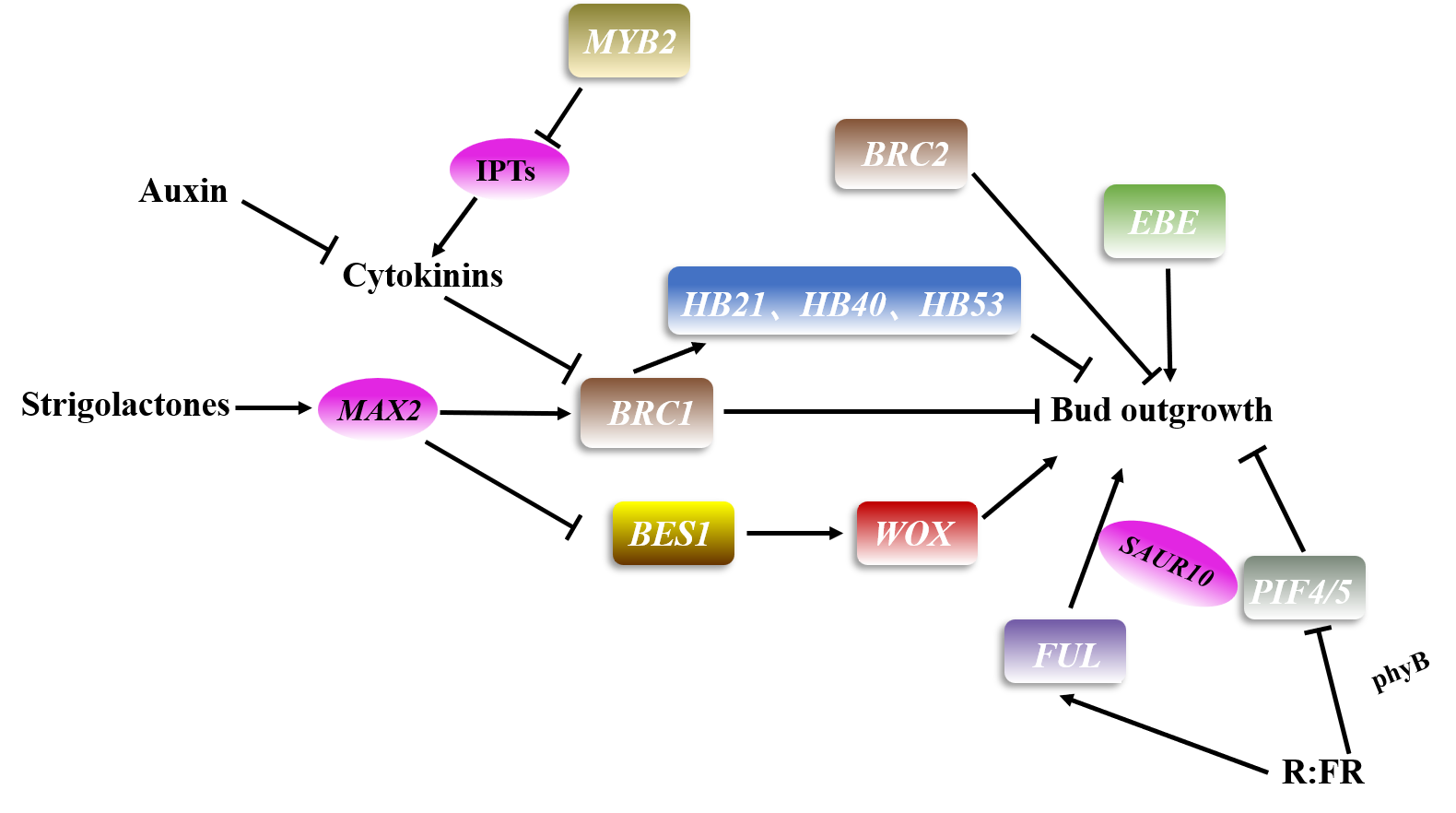
2. Axillary Meristem Outgrowth
References
- Yan, K.; Li, C.C.; Wang, Y.; Wang, X.Q.; Wang, Z.M.; Wei, D.Y.; Tang, Q.L. AGL18-1 delays flowering time through affecting expression of flowering-related genes in Brassica juncea. Plant Biotechnol. 2018, 35, 357–363.
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374.
- Groover, A.T.; Mansfield, S.D.; DiFazio, S.P.; Dupper, G.; Fontana, J.R.; Millar, R.; Wang, Y. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 2006, 61, 917–932.
- Jia, T.; Zhang, K.; Li, F.; Huang, Y.; Fan, M.; Huang, T. The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep. 2020, 39, 1755–1765.
- Lee, D.Y.; An, G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012, 69, 445–461.
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755.
- Ding, L.; Yan, S.; Jiang, L.; Liu, M.; Zhang, J.; Zhao, J.; Zhao, W.; Han, Y.Y.; Wang, Q.; Zhang, X. HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus L.). J. Exp. Bot. 2015, 66, 7075–7087.
- Lee, D.K.; Geisler, M.; Springer, P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 2009, 136, 2423–2432.
- Ikezaki, M.; Kojima, M.; Sakakibara, H.; Kojima, S.; Ueno, Y.; Machida, C.; Machida, Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010, 61, 70–82.
- Teichmann, T.; Muhr, M. Shaping plant architecture. Front. Plant Sci. 2015, 6, 233.
- Zhang, C.; Wang, J.; Wenkel, S.; Chandler, J.W.; Werr, W.; Jiao, Y. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 2018, 145, dev158352.
- Tsuda, K.; Abraham-Juarez, M.J.; Maeno, A.; Dong, Z.; Aromdee, D.; Meeley, R.; Shiroishi, T.; Nonomura, K.I.; Hake, S. KNOTTED1 Cofactors, BLH12 and BLH14, Regulate Internode Patterning and Vein Anastomosis in Maize. Plant Cell 2017, 29, 1105–1118.
- Schmitz, G.; Tillmann, E.; Carriero, F.; Fiore, C.; Cellini, F.; Theres, K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 2002, 99, 1064–1069.
- Guo, D.; Zhang, J.; Wang, X.; Han, X.; Wei, B.; Wang, J.; Li, B.; Yu, H.; Huang, Q.; Gu, H.; et al. The WRKY Transcription Factor WRKY71/EXB1 Controls Shoot Branching by Transcriptionally Regulating RAX Genes in Arabidopsis. Plant Cell 2015, 27, 3112–3127.
- Muller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597.
- Chandler, J.W.; Cole, M.; Flier, A.; Grewe, B.; Werr, W. The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 2007, 134, 1653–1662.
- Fambrini, M.; Salvini, M.; Pugliesi, C. Molecular cloning, phylogenetic analysis, and expression patterns of LATERAL SUPPRESSOR-LIKE and REGULATOR OF AXILLARY MERISTEM FORMATION-LIKE genes in sunflower (Helianthus annuus L.). Dev. Genes Evol. 2017, 227, 159–170.
- Keller, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006, 18, 598–611.
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018, 37, 1049–1060.
- Yang, F.; Wang, Q.; Schmitz, G.; Muller, D.; Theres, K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012, 71, 61–70.
- Li, Y.; Xia, T.; Gao, F.; Li, Y. Control of Plant Branching by the CUC2/CUC3-DA1-UBP15 Regulatory Module. Plant Cell 2020, 32, 1919–1932.
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957.
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76.
- Wai, A.H.; An, G. Axillary meristem initiation and bud growth in rice. J. Plant Biol. 2017, 60, 440–451.
- Katayama, N.; Tanaka, R.; Fujinami, R.; Imaichi, R. Expression pattern of CUC3 ortholog in Zeylanidium tailichenoides (Podostemaceae) infers organization of a unique distichous shoot in Podostemoideae. J. Plant Res. 2019, 132, 521–529.
- Deshpande, G.M.; Ramakrishna, K.; Chongloi, G.L.; Vijayraghavan, U. Functions for rice RFL in vegetative axillary meristem specification and outgrowth. J. Exp. Bot. 2015, 66, 2773–2784.
- Rao, N.N.; Prasad, K.; Kumar, P.R.; Vijayraghavan, U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. USA 2008, 105, 3646–3651.
- Guo, D.; Qin, G. EXB1/WRKY71 transcription factor regulates both shoot branching and responses to abiotic stresses. Plant Signal Behav. 2016, 11, e1150404.
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206.
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245.
- Muller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212.
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221.
- Yu, S.; Ligang, C.; Liping, Z.; Diqiu, Y. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 2010, 35, 459–471.
- Huang, X.; Effgen, S.; Meyer, R.C.; Theres, K.; Koornneef, M. Epistatic natural allelic variation reveals a function of AGAMOUS-LIKE6 in axillary bud formation in Arabidopsis. Plant Cell 2012, 24, 2364–2379.
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504.
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184.
- Ohmori, Y.; Tanaka, W.; Kojima, M.; Sakakibara, H.; Hirano, H.Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 2013, 25, 229–241.
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287.
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78.
- Guo, Y.; Gan, S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619.
- Bai, X.; Huang, Y.; Mao, D.; Wen, M.; Zhang, L.; Xing, Y. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 2016, 6, 19022.
- Komatsu, K.; Maekawa, M.; Ujiie, S.; Satake, Y.; Furutani, I.; Okamoto, H.; Shimamoto, K.; Kyozuka, J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 2003, 100, 11765–11770.
- Gallavotti, A.; Zhao, Q.; Kyozuka, J.; Meeley, R.; Ritter, M.; Doebley, J.; Pe, M.; Schmidt, R. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 2004, 432, 630–635.
- Meng, Q.; Li, X.; Zhu, W.; Yang, L.; Liang, W.; Dreni, L.; Zhang, D. Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis. J. Integr. Plant Biol. 2017, 59, 693–707.
- Li, C. Toward understanding the stem-cell origin and molecular regulation of rice tillering. J. Genet. Genom. 2015, 42, 47–48.
- Xu, C.; Wang, Y.; Yu, Y.; Duan, J.; Liao, Z.; Xiong, G.; Meng, X.; Liu, G.; Qian, Q.; Li, J. Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat. Commun. 2012, 3, 750.
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621.
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187.
- Min, Z.; Chen, L.; Zhang, Y.; Li, Z.Y.; Liu, M.; Li, W.P.; Ju, Y.L.; Fang, Y.L. VvBRC inhibits shoot branching in grapevine. Sci. Hortic. 2021, 289, 110370.
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296.
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468.
- Tsuji, H.; Tachibana, C.; Tamaki, S.; Taoka, K.; Kyozuka, J.; Shimamoto, K. Hd3a promotes lateral branching in rice. Plant J. 2015, 82, 256–266.
- Mashiguchi, K.; Sasaki, E.; Shimada, Y.; Nagae, M.; Ueno, K.; Nakano, T.; Yoneyama, K.; Suzuki, Y.; Asami, T. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 2009, 73, 2460–2465.
- Gao, R.; Gruber, M.Y.; Amyot, L.; Hannoufa, A. SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Mol. Biol. 2018, 96, 119–133.
- Hu, J.; Hu, X.; Yang, Y.; He, C.; Hu, J.; Wang, X. Strigolactone signaling regulates cambial activity through repression of WOX4 by transcription factor BES1. Plant Physiol. 2022, 188, 255–267.
- Bemer, M.; van Mourik, H.; Muino, J.M.; Ferrandiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403.
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790.
- Holalu, S.V.; Reddy, S.K.; Blackman, B.K.; Finlayson, S.A. Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environ. 2020, 43, 2224–2238.
- Mehrnia, M.; Balazadeh, S.; Zanor, M.I.; Mueller-Roeber, B. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013, 162, 842–857.
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520.
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566.
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010, 51, 1127–1135.




