Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Horacio Bach | -- | 2229 | 2022-08-14 07:46:26 | | | |
| 2 | Vivi Li | Meta information modification | 2229 | 2022-08-15 04:24:19 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mejía-Méndez, J.L.; Vazquez-Duhalt, R.; Hernández, L.R.; Sánchez-Arreola, E.; Bach, H. Virus-like Particles. Encyclopedia. Available online: https://encyclopedia.pub/entry/26123 (accessed on 06 March 2026).
Mejía-Méndez JL, Vazquez-Duhalt R, Hernández LR, Sánchez-Arreola E, Bach H. Virus-like Particles. Encyclopedia. Available at: https://encyclopedia.pub/entry/26123. Accessed March 06, 2026.
Mejía-Méndez, Jorge L., Rafael Vazquez-Duhalt, Luis R. Hernández, Eugenio Sánchez-Arreola, Horacio Bach. "Virus-like Particles" Encyclopedia, https://encyclopedia.pub/entry/26123 (accessed March 06, 2026).
Mejía-Méndez, J.L., Vazquez-Duhalt, R., Hernández, L.R., Sánchez-Arreola, E., & Bach, H. (2022, August 14). Virus-like Particles. In Encyclopedia. https://encyclopedia.pub/entry/26123
Mejía-Méndez, Jorge L., et al. "Virus-like Particles." Encyclopedia. Web. 14 August, 2022.
Copy Citation
Nanotechnology is a fast-evolving field focused on fabricating nanoscale objects for industrial, cosmetic, and therapeutic applications. Virus-like particles (VLPs) are self-assembled nanoparticles whose intrinsic properties, such as heterogeneity, and highly ordered structural organization are exploited to prepare vaccines; imaging agents; construct nanobioreactors; cancer treatment approaches; or deliver drugs, genes, and enzymes. However, depending upon the intrinsic features of the native virus from which they are produced, the therapeutic performance of VLPs can vary.
nanomedicine
nanotechnology
virus-like particles
1. Introduction
Nanotechnology is an interdisciplinary field devoted to engineering and developing structures ranging from 1 to 500 nm. Structures that correspond to this scale are defined as nanoparticles (NPs). NPs possess promising optical, chemical, and physical properties attractive for biomedical purposes, such as diagnostic, chemical sensing, cellular imaging, drug delivery, therapeutics, and tissue engineering [1].
Given the unique physical, optical, chemical, and therapeutic properties of NPs, there has been an increasing interest in designing methods to develop and characterize them. NPs are prepared through top-down and bottom-up approaches, including laser ablation, sputtering, etching, and mechanical milling techniques. The latter encompasses spinning, chemical reduction, molecular condensation, and green synthesis processes [2]. Instead of reducing a bulk material into nanometric objects, bottom-up methods stimulate the self-assembly of atoms into bioactive NPs. For example, in nanomedicine, the amino acid side chains and functional groups of distinct proteins (e.g., collagen, elastin, gelatin, keratin, silk, and zein) are used as scaffolds to induce the self-assembly of nanofibers, nanotubes, and nanobelts to deliver drugs or develop materials for tissue engineering [3][4].
Among protein-based nanomaterials, virus-like particles (VLPs) are self-assembled platforms commercially approved by the US Food and Drug Administration (FDA) since the 1980s [5]. Since VLPs resemble the capsid morphology, structural organization, and cellular tropism of wild-type viruses [6][7], they have been exploited to prepare monodisperse nanocarriers (20–500 nm) for drug delivery, enzyme delivery for enzymatic replacement therapy [8][9], and gene therapy applications. In addition, they have been widely used to construct human vaccines against various pathogenic viruses (e.g., human papillomavirus and zika virus) and distinct types of cancer, such as colorectal, pancreatic, and cervical cancer [5][10].
In contrast to other organic NPs, VLPs are convenient, because they exhibit higher biocompatibility, the capacity for cell internalization, and ease of functionalization for cell targeting. In fact, for the latter, they can be tailored by chemical or genetic methods with various biomolecules (i.e., transferrin, folic acid, and single-chain antibodies) to enhance their bioavailability and, herein, evoke both humoral and cellular immune responses [11][12]. However, these phenomena rely on the physicochemical and biochemical characteristics of VLPs.
Nowadays, there is a constant effort to develop analytical methods that, alone or in combination with others, enable scientists to assess the influence of physicochemical parameters in the biological activities of nanomaterials. For instance, the therapeutic performance, stability, and morphology [13][14] of VLPs can vary in accord with pH ranges [15], choice of the expression system [16][17], and purification procedures [18][19].
Since the novel coronavirus SARS-CoV-2, there has been an increasing interest in reviewing VLPs as a powerful approach to producing vaccines and nanocarriers [20]. However, there is a need to complement those studies and recent ones [21], with basic principles about expressing, manipulating, functionalizing, and characterizing VLPs.
Therefore, the literature regarding the structure classification, production, morphology, and functionalization of VLPs for biomedical applications was consulted in this entry. The research engines PubMed, Google Scholar, Web of Science, and Wiley Online Library databases were used to compile the literature. In addition, the same databases were used to integrate the main aspects of viruses and how the geometry is indispensable to constructing VLPs-based platforms.
2. Brief Description of Viruses
Viruses are entities characterized by the lack of machinery for self-replication and energy production. Their replication relies on hijacking the cellular systems of the host cells to produce the molecules necessary for their assembly and subsequent escape from the cells. Infectious viruses that have not been internalized in the host cells (not in a replication phase) and residing outside the cell host are called virions.
Viruses are ubiquitous entities containing a DNA- or RNA-based genome protected or not by a protein shell known as a capsid and other accessory biomolecules, such as proteins and membranes [22]. The capsid shell is assembled through covalent and electrostatic interactions conferring a robust and flexible structure and made by small subunits known as capsomers [23]. These capsomers are critical components of the VLPs and constitute the basis for their self-assembly into complex structures [24].
The general structures of viral capsids show a diversity of conformations, including icosahedral conformation. Icosahedral capsids comprise 20 triangular subunits, whereas helical capsids are proteins assembled to form helical cylinders [25]. Despite the structural variabilities between both shapes, the functions of the viral capsid rely on the packaging, sequestering, and protection of the viral genetic material, preventing its degradation in the environment or exposure to chemical hazards [25][26].
Besides the capsids, viruses are surrounded or not by a viral envelope that facilitates their fusion and infection of the host cell. In addition to the presence of proteins and glycoproteins, this envelope can contain lipidic membranes acquired from the host. Viral envelopes are necessary to protect the viral genome, increase the packaging capacity, confer structural flexibility, and enable the new viruses to exit from their host cell and avert immune responses [27].
Viruses with envelopes are known as enveloped viruses, such as the varicella-zoster virus [28], lymphocytic choriomeningitis virus, tick-borne encephalitis virus, human immunodeficiency virus 1 [29], and severe acute respiratory syndrome coronavirus [30][31].
3. Key Concepts about Virus-like Particles (VLPs)
The assembly of VLPs uses exclusively viral proteins and excludes the genetic material. Thus, VLPs can be produced from a myriad of wild-type viruses, such as hepatitis B and E, tobacco, and papaya mosaic viruses [32], and from the capsids of various bacteriophages (Qβ, MS2, P22, and PP7) [33][34][35][36].
The production of VLPs using bacteria, mammalian cells, plants, yeast, and insect cell lines is well-documented [37][38][39]. However, current efforts are devoted to understanding their capsid self-assembly mechanisms to improve the cargo loading capacity, potency, and efficacy.
The capsid self-assembly of VLPs is a spontaneous natural process by which highly ordered structures arise from the interactions between protein monomers, also known as building blocks. The self-association between building blocks is facilitated by a thermodynamic equilibrium based on van der Waals, hydrogen bonding, hydrophobic, and electrostatic interactions during the nucleation and growth phases [40][41]. As a result, VLPs can adopt different structural arrangements, such as helical, icosahedral, spherical, or complex shapes [42][43] (Figure 1).
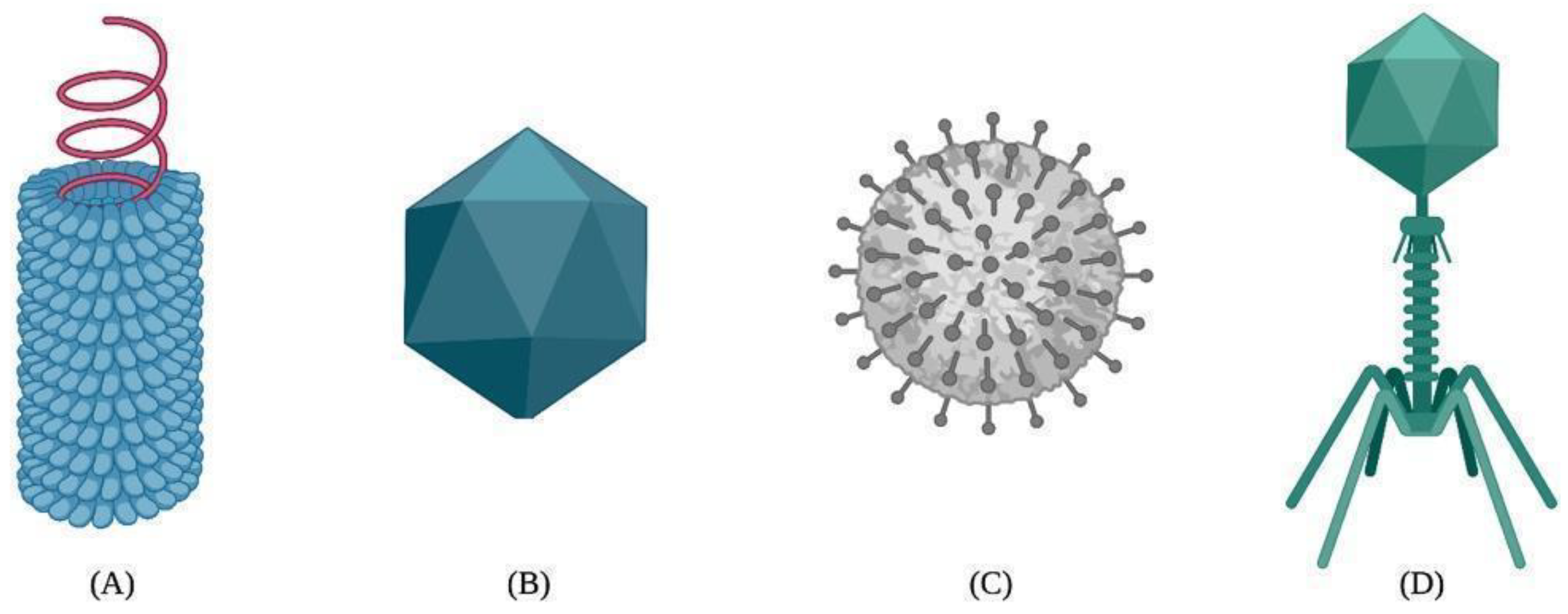
Figure 1. Different types of viral capsids: (A) helical, (B) icosahedral, (C) spherical, and (D) complex.
The self-assembly of VLPs is assisted by small molecules such as scaffolding proteins and nucleic acids [40]. In this regard, scaffolding proteins and nucleic acids can be used to aid the in vitro capsid self-assembly process of many VLPs, such as the cowpea chlorotic mottle virus, hepatitis C virus, bacteriophage MS2, simian virus 40, beak and feather disease virus, and adeno-associated virus serotype 2 VLPs [40].
Since the self-assembly of VLPs can be perturbed by changes in the salt concentration, denaturant agents, pH, and temperature [28], excipients such as polysorbate 80 are used to avoid the aggregation and preserve the stability of the VLPs [40]. On the other hand, molecules such as 2-phenoxyethanol have been used as a preservative agent to produce licensed VLP-based vaccines such as the Engerix®-B hepatitis B vaccine [44]. Comparably, buffering agents such as L-histidine and sodium borate have been used to manufacture the Gardasil® human papillomavirus vaccine [44].
VLP-based nanocarriers could be produced by producing viruses in their natural host, followed by virion purification, disassembling, and nucleic acid removal (see Figure 2). Then, the disassembled coat proteins are transferred into assembling conditions in the presence of any cargo to be encapsulated. Finally, the VLPs containing the load could be functionalized with any ligand or chemical moiety and covered with polymers or other compounds to reduce or enhance their immunogenicity or facilitate cell internalization. Molecules used to decorate the VLP surface include T-cell receptor ligands; polysaccharides; enzymes (e.g., α-glucosidase); and canonical amino acids (i.e., aspartic acid, cysteine, glutamic acid, lysine, and tyrosine); among others [45][46][47][48].
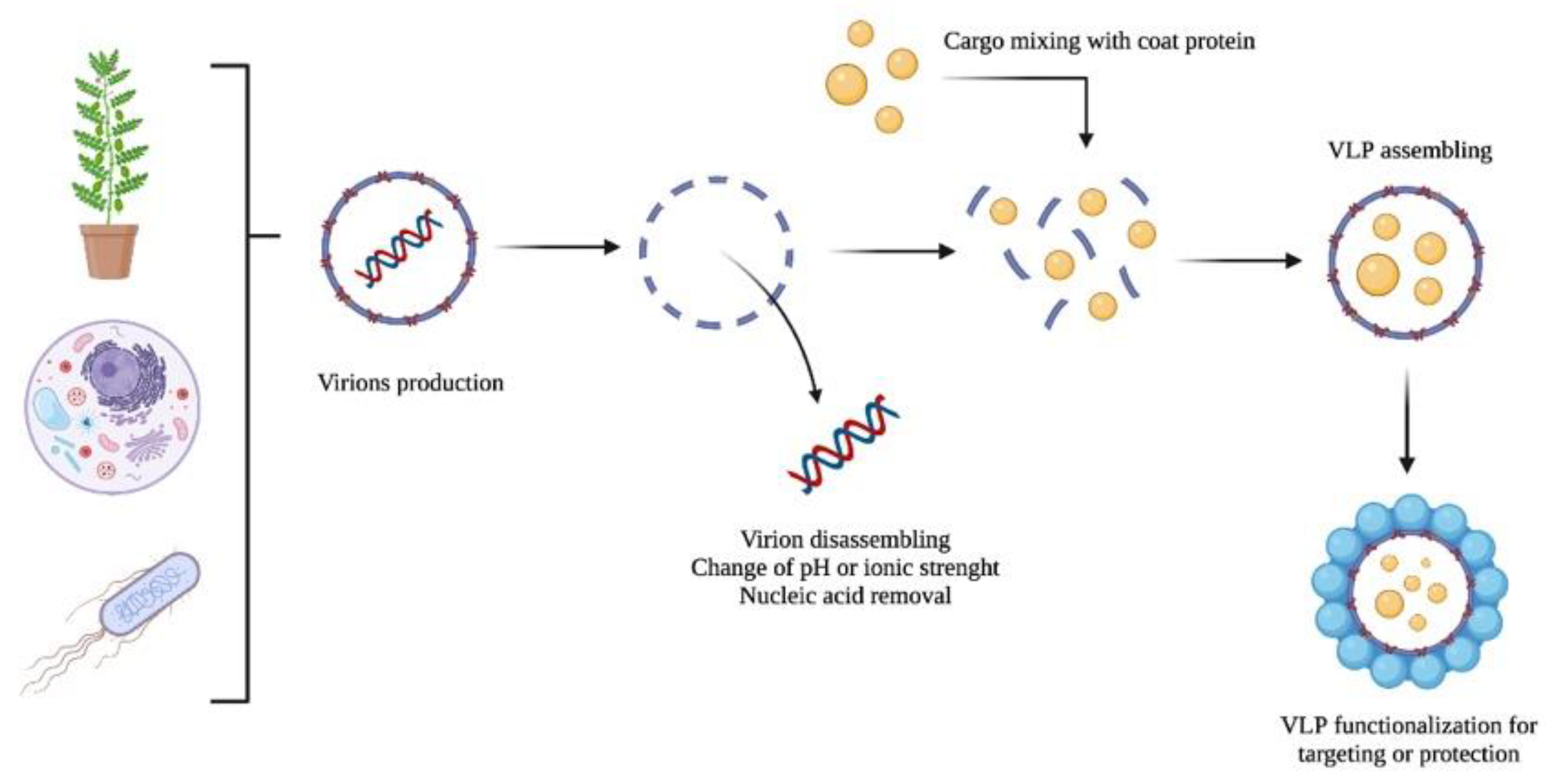
Figure 2. A schematic representation of the VLP production of virions as nanocarriers: (i) production, (ii) disassembling and nucleic acid removal, (iii) cargo encapsulation, and (iv) VLP functionalization.
The coat proteins of the VLPs can be heterologously produced and purified from an industrial microorganism and reconstituted under suitable conditions in the presence of their cargo. In this regard, the cargo loading could be obtained by simple encapsulation when the charges of the inner portion of the VLP and the cargo surface are complementary. However, as described in detail below, the loading could also be reached by chemical or genetic modification of the coat protein on both the inner and outer surfaces.
Although most VLPs are deficient in viral genetic material, they retain properties from the native virus from which they are produced. These features include their affinity for cellular receptors, host cell entry mechanisms, and immunogenicity [49]. As native viruses, VLPs elicit both humoral and cellular responses in the host system. These responses are stimulated by VLP-based systems due to their repetitive antigenic epitopes, resembling pathogen-associated molecular patterns (PAMPs), and low polydispersity [50]. The introduction of antigens (e.g., proteins or small peptides) on the VLP surface is necessary to enhance the immune response and is achieved through chemical and genetic approaches [51][52][53]. It is worth mentioning that the immune response only occurs against the antigen, not the rest of the nanostructure produced [54].
Functionalized VLPs can trigger an immune response by activating PAMPs. These patterns are conserved molecular motifs associated with pathogen infections [55]. PAMPs are recognized by pattern–recognition receptors (PRRs), nucleotide-binding oligomerization domain-like receptors, and Toll-like receptors (TLRs) on the surface of phagocytic cells [56]. Due to the similarity of VLPs with wild-type viruses, the morphology of these NPs stimulates adaptive and innate immune responses, including cellular uptake. For example, VLPs can be taken up by enterocytes, specialized intestinal epithelial cells, dendritic cells (DCs), and macrophages [57]. This process depends upon the size, shape, and surface charge of the VLPs [58].
Like other NPs, the surfaces of VLPs can be modified with various ligands to improve their therapeutic efficacy, bioavailability, and cellular interactions. The introduction of multiple molecules on the VLPs surface is known as multivalence. Then, VLPs are multivalent engineered nanoplatforms that form independent ligand–receptor bonds, elicit therapeutic responses, and mitigate endemic diseases. For example, Garg and coworkers developed a VLP-based multivalent vaccine (CJaYZ) against four arboviruses, including the zika, chikungunya, yellow fever, and Japanese encephalitis viruses. Even though the CjaYZ vaccine promoted a neutralizing antibody response against the four viruses in Balb/c mice models, further studies are required to assess the precise antigen amount of individual VLPs and their efficacy in other animal models [59]. The general features of VLPs are depicted in Figure 3.
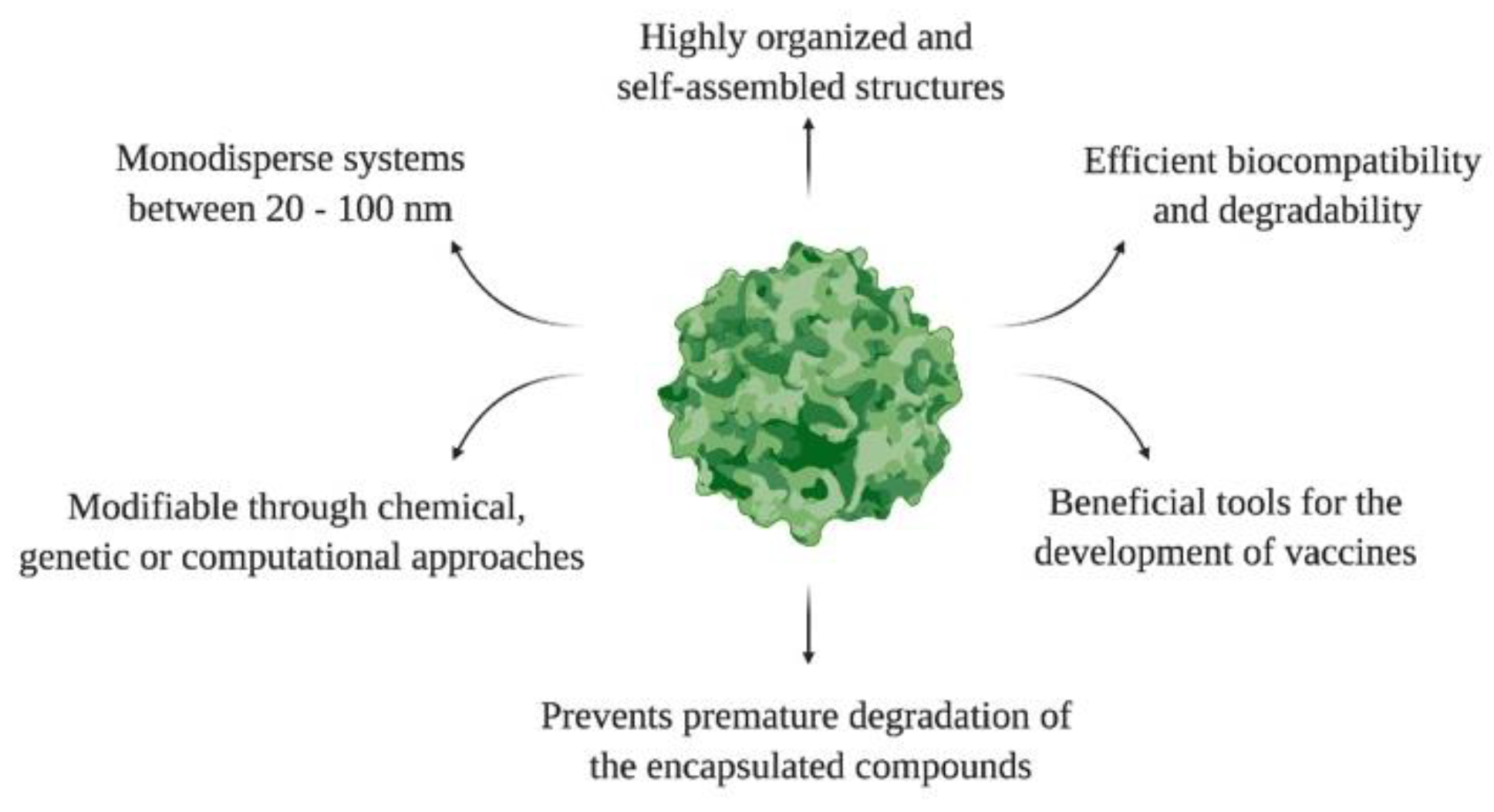
Figure 3. General features of VLPs.
4. Structure Classification of VLPs
Given the structural diversity of VLPs, they have been categorized into three main groups: enveloped, nonenveloped, and chimeric. For example, enveloped VLPs (eVLPs) are expressed using eukaryotic systems and as wild-type viruses. eVLPs are complex structures that own a host–cell-derived membrane and one or more glycoproteins. The viral envelope in eVLPs can be engineered to display heterologous adjuvants and antigens; however, this process might alter their downstream processing due to the possible presence of host cellular contaminants [60].
As vaccines, eVLPs stimulate immune responses and are manipulated with chemical or genetic methods. In this regard, a handful of eVLPs have been produced from pathogenic viruses for vaccine development. Some examples include eVLPs derived from the West Nile virus (WNV), dengue virus (DENV), JEV, Rift Valley fever virus (RVFV), and Ross River virus (RRV), among others [61][62]. For drug delivery applications, Rous sarcoma virus (RSV) eVLPs displaying a single-chain variable fragment (scFv) of humanized CC49 antibody (hCC49) have been expressed on silkworm larvae to deliver doxorubicin into human colon carcinoma cells [63]. In addition, doxorubicin was loaded into hCC49 scFv-displaying RSV VLPs by electroporation.
Non-eVLPs are single or multiple capsid protein nanoconstructs that lack cell membranes. Members of this category are produced on eukaryotic or prokaryotic expression systems. The surfaces of non-eVLPs can also be manipulated with chemical and genetic approaches to display epitopes or peptides on their surfaces and, herein, elicit wider immunological responses [64]. For instance, non-eVLPs derived from the coxsackievirus B3 antigen have enhanced humoral immune responses and protected murine models against myocarditis [65]. Additionally, rotavirus non-eVLPs (Ro-VLPs) were produced in Nicotiana benthamiana plants. The immunogenicity and tolerance of Ro-VLPs were evaluated in adults, toddlers, and infants [66].
Chimeric VLPs (cVLPs) are nanoplatforms from structural components originating from at least two different viral serotypes [67]. In these nanoplatforms, the VLP core can be modified with antigens that cannot self-assemble or present polyproteins from distinct viruses [68]. In contrast with the other two categories of VLPs, cVLPs are useful to present foreign epitopes; entrap multiple therapeutic or diagnostic molecules; and target cells, tissues, or organs [42]. However, the production of cVLPs depends upon various factors, such as the type of conjugation between proteins, glycosylation patterns, cell type, length of the fused antigen, and steric effects.
For biomedical purposes, cVLPs have been prepared from major structural components of influenza viruses (e.g., M1 protein) and human immunodeficiency virus type-1 (HIV-1) (e.g., Group-specific antigen; Gag) to target colon carcinoma cell lines and for vaccination purposes [69]. In another study, murine polyomavirus cVLPs were manipulated to elicit CD8+ and CD4+ T cells and antibody responses [70]. The immune response against other cVLPs has also been evaluated recently, specifically for the foot-and-mouth disease virus cVLP vaccine, based on the co-expression of the HIV-1 Gag protein and rabies glycoproteins [71].
References
- Murphy, C.J.; Vartanian, A.M.; Geiger, F.M.; Hamers, R.J.; Pedersen, J.; Cui, Q.; Haynes, C.L.; Carlson, E.E.; Hernandez, R.; Klaper, R.D.; et al. Biological Responses to Engineered Nanomaterials: Needs for the Next Decade. ACS Cent. Sci. 2015, 1, 117–123.
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A Review of Synthesis Methods, Properties, Recent Progress, and Challenges. Mater. Adv. 2021, 2, 1821–1871.
- DeFrates, K.G.; Moore, R.; Borgesi, J.; Lin, G.; Mulderig, T.; Beachley, V.; Hu, X. Protein-Based Fiber Materials in Medicine: A Review. Nanomaterials 2018, 8, 457.
- Mandal, D.; Nasrolahi Shirazi, A.; Parang, K. Self-Assembly of Peptides to Nanostructures. Org. Biomol. Chem. 2014, 12, 3544–3561.
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front. Microbiol. 2022, 12, 790121.
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines against Respiratory Viruses. Front. Immunol. 2019, 10, 22.
- Jhaveri, J.; Raichura, Z.; Khan, T.; Momin, M.; Omri, A. Chitosan Nanoparticles-Insight into Properties, Functionalization and Applications in Drug Delivery and Theranostics. Molecules 2021, 26, 272.
- Gama, P.; Cadena-Nava, R.D.; Juarez-Moreno, K.; Pérez-Robles, J.; Vazquez-Duhalt, R. Virus-Based Nanoreactors with GALT Activity for Classic Galactosemia Therapy. ChemMedChem 2021, 16, 1438–1445.
- Wang, Y.; Douglas, T. Protein Nanocage Architectures for the Delivery of Therapeutic Proteins. Curr. Opin. Colloid Interface Sci. 2021, 51, 101395.
- Mohsen, M.O.; Speiser, D.E.; Knuth, A.; Bachmann, M.F. Virus-like Particles for Vaccination against Cancer. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1579.
- Cho, K.; Wang, X.; Nie, S.; Chen, Z.; Shin, D.M. Therapeutic Nanoparticles for Drug Delivery in Cancer. Clin. Cancer Res. 2008, 14, 1310–1316.
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) from Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100.
- Ding, X.; Liu, D.; Booth, G.; Gao, W.; Lu, Y. Virus-Like Particle Engineering: From Rational Design to Versatile Applications. Biotechnol. J. 2018, 13, 1700324.
- Wang, Y.; Wang, G.; Duan, W.-T.; Sun, M.-X.; Wang, M.-H.; Wang, S.-H.; Cai, X.-H.; Tu, Y. Self-Assembly into Virus–like Particles of the Recombinant Capsid Protein of Porcine Circovirus Type 3 and Its Application on Antibodies Detection. AMB Expr. 2020, 10, 3.
- Wilkerson, J.W.; Yang, S.-O.; Funk, P.J.; Stanley, S.K.; Bundy, B.C. Nanoreactors: Strategies to Encapsulate Enzyme Biocatalysts in Virus-like Particles. New Biotechnol. 2018, 44, 59–63.
- Donaldson, B.; Lateef, Z.; Walker, G.F.; Young, S.L.; Ward, V.K. Virus-like Particle Vaccines: Immunology and Formulation for Clinical Translation. Expert Rev. Vaccines 2018, 17, 833–849.
- Qian, C.; Liu, X.; Xu, Q.; Wang, Z.; Chen, J.; Li, T.; Zheng, Q.; Yu, H.; Gu, Y.; Li, S.; et al. Recent Progress on the Versatility of Virus-Like Particles. Vaccines 2020, 8, 139.
- Cervera, L.; Gòdia, F.; Tarrés-Freixas, F.; Aguilar-Gurrieri, C.; Carrillo, J.; Blanco, J.; Gutiérrez-Granados, S. Production of HIV-1-Based Virus-like Particles for Vaccination: Achievements and Limits. Appl. Microbiol. Biotechnol. 2019, 103, 7367–7384.
- Carvalho, S.B.; Silva, R.J.S.; Moreira, A.S.; Cunha, B.; Clemente, J.J.; Alves, P.M.; Carrondo, M.J.T.; Xenopoulos, A.; Peixoto, C. Efficient Filtration Strategies for the Clarification of Influenza Virus-like Particles Derived from Insect Cells. Sep. Purif. Technol. 2019, 218, 81–88.
- Lu, W.; Zhao, Z.; Huang, Y.-W.; Wang, B. Review: A Systematic Review of Virus-like Particles of Coronavirus: Assembly, Generation, Chimerism and Their Application in Basic Research and in the Clinic. Int. J. Biol. Macromol. 2022, 200, 487–497.
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59.
- Wilusz, J. The Fundamentals of Human Virology. In Microbial Forensics; Academic Press: Cambridge, MA, USA, 2005; pp. 41–53. Available online: https://reader.elsevier.com/reader/sd/pii/B9780120884834500068?token=D05E2ABB6D7A53A0599AEC3B4DA2198639C8B92C5885FD1DAD03FD3B815E2AD22A5A5B7DA37908EDC614D5F34F18C331&originRegion=us-east-1&originCreation=20220418192715 (accessed on 18 April 2022).
- Louten, J. Chapter 2—Virus Structure and Classification. In Essential Human Virology; Academic Press: Cambridge, MA, USA, 2016; pp. 19–29. Available online: https://reader.elsevier.com/reader/sd/pii/B9780128009475000028?token=7BE372B98B22B5FE56B2BC5017996E8FFCD5C89A6B07CDF8E31722FCCE498BAAB6E0E964180573473995CB6AC8418EE3&originRegion=us-east-1&originCreation=20220418192735 (accessed on 18 April 2022).
- Lamarre, B.; Ryadnov, M.G. Self-Assembling Viral Mimetics: One Long Journey with Short Steps. Macromol. Biosci. 2011, 11, 503–513.
- Pellett, P.E.; Mitra, S.; Holland, T.C. Basics of Virology. In Handbook of Clinical Neurology; Academic Press: Cambridge, MA, USA, 2014; pp. 45–66. Available online: https://reader.elsevier.com/reader/sd/pii/B978044453488000002X?token=8867B3E02B07251123A161859F00BE6AD3C8B28CADBE87515D622735E64D2280DD3A91ACEE30FBEA7FDDAA869EC19B6C&originRegion=us-east-1&originCreation=20220418192824 (accessed on 18 April 2022).
- Hellen, C.U.T.; Wimmer, E. The Role of Proteolytic Processing in the Morphogenesis of Virus Particles. Experientia 1992, 48, 201–215.
- Wisskirchen, K.; Lucifora, J.; Michler, T.; Protzer, U. New Pharmacological Strategies to Fight Enveloped Viruses. Trends Pharmacol. Sci. 2014, 35, 470–478.
- Lan, K.; Luo, M.-H. Herpesviruses: Epidemiology, Pathogenesis, and Interventions. Virol. Sin. 2017, 32, 347–348.
- Fenner, F.; Bachmann, P.A.; Gibbs, E.P.J.; Murphy, F.A.; Studdert, M.J.; White, O.D. Structure and Composition of Viruses. In Veterinary Virology; Academic Press: Cambridge, MA, USA, 1987; pp. 3–19. Available online: https://reader.elsevier.com/reader/sd/pii/B9780122530555500050?token=839CB2DC605C5E4B8DDE945EEDC256BFFC9A0F47195CEEFD3E738A406FE0AD93976DB1DFC83C9863DE3C92ABD4B031B7&originRegion=us-east-1&originCreation=20220418192957 (accessed on 18 April 2022).
- Schoeman, D.; Fielding, B.C. Coronavirus Envelope Protein: Current Knowledge. Virol. J. 2019, 16, 69.
- Cosset, F.-L.; Lavillette, D. 4—Cell Entry of Enveloped Viruses. In Advances in Genetics; Academic Press: Cambridge, MA, USA, 2011; pp. 121–183. Available online: https://reader.elsevier.com/reader/sd/pii/B9780123808608000045?token=5B7798F78728BD7418D95FAAFA97024C551D30E63F8BA3CA8B48A665E51F5D514C184E7B0F81D10B46C8A2A3A21D35B7&originRegion=us-east-1&originCreation=20220418193042 (accessed on 18 April 2022).
- López-Macías, C. Virus-like Particle (VLP)-Based Vaccines for Pandemic Influenza: Performance of a VLP Vaccine during the 2009 Influenza Pandemic. Hum. Vaccines Immunother. 2012, 8, 411–414.
- Cui, Z.; Gorzelnik, K.V.; Chang, J.-Y.; Langlais, C.; Jakana, J.; Young, R.; Zhang, J. Structures of Qβ Virions, Virus-like Particles, and the Qβ–MurA Complex Reveal Internal Coat Proteins and the Mechanism of Host Lysis. Proc. Natl. Acad. Sci. USA 2017, 114, 11697–11702.
- Golmohammadi, R.; Valegård, K.; Fridborg, K.; Liljas, L. The Refined Structure of Bacteriophage MS2 at 2·8 Å Resolution. J. Mol. Biol. 1993, 234, 620–639.
- Herbert, F.C.; Brohlin, O.R.; Galbraith, T.; Benjamin, C.; Reyes, C.A.; Luzuriaga, M.A.; Shahrivarkevishahi, A.; Gassensmith, J.J. Supramolecular Encapsulation of Small-Ultrared Fluorescent Proteins in Virus-Like Nanoparticles for Noninvasive In Vivo Imaging Agents. Bioconj. Chem. 2020, 31, 1529–1536.
- Rohovie, M.J.; Nagasawa, M.; Swartz, J.R. Virus-like Particles: Next-Generation Nanoparticles for Targeted Therapeutic Delivery. Bioeng. Transl. Med. 2017, 2, 43–57.
- Syomin, B.V.; Ilyin, Y.V. Virus-Like Particles as an Instrument of Vaccine Production. Mol. Biol. 2019, 53, 323–334.
- Bárcena, J.; Blanco, E. Design of Novel Vaccines Based on Virus-Like Particles or Chimeric Virions. In Structure and Physics of Viruses: An Integrated Textbook; Subcellular Biochemistry; Mateu, M.G., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 631–665. ISBN 978-94-007-6552-8.
- Shirbaghaee, Z.; Bolhassani, A. Different Applications of Virus-like Particles in Biology and Medicine: Vaccination and Delivery Systems. Biopolymers 2016, 105, 113–132.
- Le, D.T.; Müller, K.M. In Vitro Assembly of Virus-Like Particles and Their Applications. Life 2021, 11, 334.
- Grzelczak, M.; Vermant, J.; Furst, E.M.; Liz-Marzán, L.M. Directed Self-Assembly of Nanoparticles. ACS Nano 2010, 4, 3591–3605.
- Pushko, P.; Pumpens, P.; Grens, E. Development of Virus-Like Particle Technology from Small Highly Symmetric to Large Complex Virus-Like Particle Structures. INT 2013, 56, 141–165.
- Singh, K.; Marasini, B.; Chen, X.; Ding, L.; Wang, J.-J.; Xiao, P.; Villinger, F.; Spearman, P. A Bivalent, Spherical Virus-Like Particle Vaccine Enhances Breadth of Immune Responses against Pathogenic Ebola Viruses in Rhesus Macaques. J. Virol. 2020, 94, e01884-19.
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering Virus-like Particles as Vaccines. Biotechnol. Bioeng. 2014, 111, 425–440.
- Derdak, S.V.; Kueng, H.J.; Leb, V.M.; Neunkirchner, A.; Schmetterer, K.G.; Bielek, E.; Majdic, O.; Knapp, W.; Seed, B.; Pickl, W.F. Direct Stimulation of T Lymphocytes by Immunosomes: Virus-like Particles Decorated with T Cell Receptor/CD3 Ligands plus Costimulatory Molecules. Proc. Natl. Acad. Sci. USA 2006, 103, 13144–13149.
- Hills, R.A.; Howarth, M. Virus-like Particles against Infectious Disease and Cancer: Guidance for the Nano-Architect. Curr. Opin. Biotechnol. 2022, 73, 346–354.
- Cayetano-Cruz, M.; Coffeen, C.F.; Valadez-García, J.; Montiel, C.; Bustos-Jaimes, I. Decoration of Virus-like Particles with an Enzymatic Activity of Biomedical Interest. Virus Res. 2018, 255, 1–9.
- Smith, M.T.; Hawes, A.K.; Bundy, B.C. Reengineering Viruses and Virus-like Particles through Chemical Functionalization Strategies. Curr. Opin. Biotechnol. 2013, 24, 620–626.
- Kawano, M.; Matsui, M.; Handa, H. Chapter 15—Technologies That Generate and Modify Virus-like Particles for Medical Diagnostic and Therapy Purposes. In Design and Development of New Nanocarriers; Grumezescu, A.M., Ed.; William Andrew Publishing: Norwich, NY, USA, 2018; pp. 555–594. ISBN 978-0-12-813627-0.
- Akdis, M.; Akdis, C.A. Therapeutic Manipulation of Immune Tolerance in Allergic Disease. Nat. Rev. Drug Discov. 2009, 8, 645–660.
- Frietze, K.M.; Peabody, D.S.; Chackerian, B. Engineering Virus-like Particles as Vaccine Platforms. Curr. Opin. Virol. 2016, 18, 44–49.
- O’Rourke, J.P.; Peabody, D.S.; Chackerian, B. Affinity Selection of Epitope-Based Vaccines Using a Bacteriophage Virus-like Particle Platform. Curr. Opin. Virol. 2015, 11, 76–82.
- Brown, S.D.; Fiedler, J.D.; Finn, M.G. Assembly of Hybrid Bacteriophage Qβ Virus-like Particles. Biochemistry 2009, 48, 11155–11157.
- Barrett, J.C.; Acar, H.; Mellas, M.J.; Tirrell, M.V. 11—Peptides in Immunoengineering. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 287–326. ISBN 978-0-08-100736-5.
- Heise, M.T. Viral Pathogenesis. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-12-801238-3.
- Kato, J.; Svensson, C.I. Chapter Nine—Role of Extracellular Damage-Associated Molecular Pattern Molecules (DAMPs) as Mediators of Persistent Pain. In Progress in Molecular Biology and Translational Science; Molecular and Cell Biology of Pain; Price, T.J., Dussor, G., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 131, pp. 251–279.
- Karandikar, S.; Mirani, A.; Waybhase, V.; Patravale, V.B.; Patankar, S. Chapter 10—Nanovaccines for Oral Delivery-Formulation Strategies and Challenges. In Nanostructures for Oral Medicine; Micro and Nano Technologies; Andronescu, E., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 263–293. ISBN 978-0-323-47720-8.
- Crisci, E.; Bárcena, J.; Montoya, M. Virus-like Particles: The New Frontier of Vaccines for Animal Viral Infections. Vet. Immunol. Immunopathol. 2012, 148, 211–225.
- Garg, H.; Mehmetoglu-Gurbuz, T.; Joshi, A. Virus Like Particles (VLP) as Multivalent Vaccine Candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Sci. Rep. 2020, 10, 4017.
- Chen, C.-W.; Saubi, N.; Joseph-Munné, J. Design Concepts of Virus-Like Particle-Based HIV-1 Vaccines. Front. Immunol. 2020, 11, 573157.
- Pijlman, G.P. Enveloped Virus-like Particles as Vaccines against Pathogenic Arboviruses. Biotechnol. J. 2015, 10, 659–670.
- Cheng, F.; Mukhopadhyay, S. Generating Enveloped Virus-like Particles with in Vitro Assembled Cores. Virology 2011, 413, 153–160.
- Kato, T.; Yui, M.; Deo, V.K.; Park, E.Y. Development of Rous Sarcoma Virus-like Particles Displaying HCC49 ScFv for Specific Targeted Drug Delivery to Human Colon Carcinoma Cells. Pharm. Res. 2015, 32, 3699–3707.
- Ong, H.K.; Tan, W.S.; Ho, K.L. Virus like Particles as a Platform for Cancer Vaccine Development. PeerJ 2017, 5, e4053.
- Zhang, L.; Parham, N.J.; Zhang, F.; Aasa-Chapman, M.; Gould, E.A.; Zhang, H. Vaccination with Coxsackievirus B3 Virus-like Particles Elicits Humoral Immune Response and Protects Mice against Myocarditis. Vaccine 2012, 30, 2301–2308.
- Kurokawa, N.; Robinson, M.K.; Bernard, C.; Kawaguchi, Y.; Koujin, Y.; Koen, A.; Madhi, S.; Polasek, T.M.; McNeal, M.; Dargis, M.; et al. Safety and Immunogenicity of a Plant-Derived Rotavirus-like Particle Vaccine in Adults, Toddlers and Infants. Vaccine 2021, 39, 5513–5523.
- Rutkowska, D.A.; Mokoena, N.B.; Tsekoa, T.L.; Dibakwane, V.S.; O’Kennedy, M.M. Plant-Produced Chimeric Virus-like Particles—A New Generation Vaccine against African Horse Sickness. BMC Vet. Res. 2019, 15, 432.
- Grgacic, E.V.L.; Anderson, D.A. Virus-like Particles: Passport to Immune Recognition. Methods 2006, 40, 60–65.
- Deo, V.K.; Kato, T.; Park, E.Y. Chimeric Virus-Like Particles Made Using GAG and M1 Capsid Proteins Providing Dual Drug Delivery and Vaccination Platform. Mol. Pharm. 2015, 12, 839–845.
- Pattinson, D.J.; Apte, S.H.; Wibowo, N.; Rivera-Hernandez, T.; Groves, P.L.; Middelberg, A.P.J.; Doolan, D.L. Chimeric Virus-Like Particles and Capsomeres Induce Similar CD8+ T Cell Responses but Differ in Capacity to Induce CD4+ T Cell Responses and Antibody Responses. Front. Immunol. 2020, 11, 564627.
- Fontana, D.; Garay, E.; Cervera, L.; Kratje, R.; Prieto, C.; Gòdia, F. Chimeric VLPs Based on HIV-1 Gag and a Fusion Rabies Glycoprotein Induce Specific Antibodies against Rabies and Foot-and-Mouth Disease Virus. Vaccines 2021, 9, 251.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Revisions:
2 times
(View History)
Update Date:
15 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





