| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Asma Hussain Alkatheri | -- | 3757 | 2022-08-10 16:35:19 | | | |
| 2 | Catherine Yang | + 1 word(s) | 3758 | 2022-08-11 12:28:20 | | |
Video Upload Options
The bacterial membrane is part of a secretion system which plays an integral role to secrete proteins responsible for cell viability and pathogenicity; pathogenic bacteria, for example, secrete virulence factors and other membrane-associated proteins to invade the host cells through various types of secretion systems (Type I to Type IX). The bacterial membrane can also mediate microbial communities’communication through quorum sensing (QS), by secreting auto-stimulants to coordinate gene expression. The process of bacterial attachment to host cells is the first step in initiating a series of biochemical reactions that include secretion of lethal substances and toxins from the bacterial cytoplasm and penetration of the host cell wall, resulting in infection establishment.
1. Introduction
The bacterial membrane is the protective barrier of the bacteria that helps transport proteins, virulence factors, solutes, and chemical signals [1]. There are various types of secretion systems in the bacterial cell that play important roles to either persist against host cells or to provide a response to the external environment. These secretion system attributes also enable the bacterial cell not only to compete with other microorganisms for host cell attachment, but also to provide immune evasion during the infection process [2].
Chemical signals are secreted through Quorum sensing (QS) in the bacterial membrane, which acts on cell-to-cell communication as an indication of population density [3][4]. Considering the important role played by the bacterial membrane in causing infections, use of antibiotics that target the bacterial membrane through different mechanisms would help in managing antimicrobial resistance [5][6][7].
Due to excessive use of existing antibiotics and the lack of new drug development, emergence of multidrug resistance (MDR) has caused a wide spread of infection worldwide, making MDR bacteria a threat to global health [8]. This mechanism of resistance may arise from antibiotic target modifications, such as the penicillin-binding proteins, and/or a change in the membrane permeability to antibiotics [9]. In addition, modification of the lipopolysaccharide charge can reduce the affinity of the antimicrobial to a membrane, causing resistance [10].
2. Bacterial Membrane Interaction with Host Cells
Virulence factors are produced within the microorganism and secreted through membranes for adherence and for host cell invasion [11]. The process of bacterial attachment to host cells is the first step in initiating a series of biochemical reactions that include secretion of lethal substances and toxins from the bacterial cytoplasm and penetration of the host cell wall, resulting in infection establishment [12]. For example, virulence factors are important for the adhesion of Escherichia coli to the epithelium of the urinary tract, making it easier for E. coli to be transported through the urethra and uterus [13].
The host’s susceptibility to bacterial infection depends on the virulence of the bacteria’s virulence factors and the effectiveness of the host’s physiological and immunological systems. Infection begins when the virulence factors overcome the prevailing balance between them and the host’s immunity [11]. In addition, some bacteria possess properties that help them escape from the host’s immune system. Most of these processes are associated with the bacterial membrane [14]. Most bacteria use secretion systems which consist of transporters located in the bacterial membrane that are concerned with transporting proteins to the external environment or directly to the cells of the host. These proteins work to adapt the bacteria from their environment, create shelter, release DNA, and attack host cells [15]. Furthermore, a system called QS helps bacteria communicate with their external environment and coordinate their biological processes by secreting chemical signals through the bacterial membrane to compete or communicate with the bacteria in the surrounding environment [15][16]. This indicates that both the secretion and QS systems are closely associated to the bacterial membrane.
2.1. Secretion Systems in the Bacteria Membrane
Bacteria rely on secretion systems to secrete proteins to compete with neighboring microorganisms for ecological niches. For instance, the secretion system of Pseudomonas aeruginosa plays a role in triggering lung infections in cystic fibrosis patients, as the high viscosity of the mucus layer and disruption of mucociliary clearance are the main reasons why they are more susceptible to infections [17].
The secretion system is found in almost all bacterial species and is divided into eight types according to structure and activity: T1SS, T2SS, T3SS, T4SS, T5SS, T6SS, T7SS, and T9SS [15]. T8SS seem to have a limited role and, as it is not sufficiently understood, it will not be discussed in the current review. T1SS, T2SS, T3SS, T5SS, T6SS and T9SS are exclusively found in Gram-negative bacteria, while T4SS and T7SS can be found in both Gram-positive and Gram-negative bacteria, which will be discussed later. All secretion systems combined are shown in Figure 1 below.
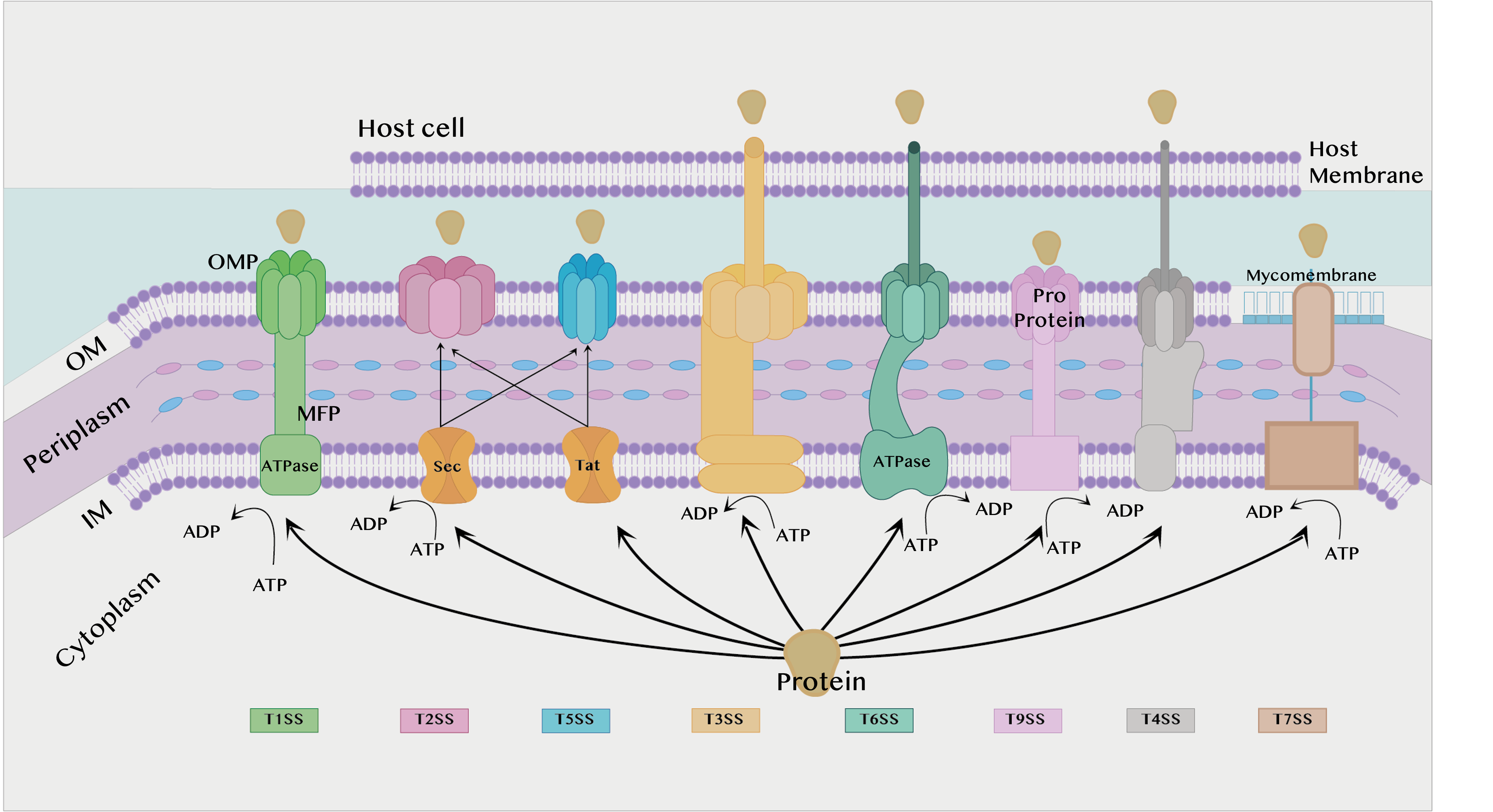
Figure 1. Different types of secretion systems. IM: Inner membrane. OM: Outer membrane. ATP: Adenosine triphosphate. ATPase: Adenosine triphosphatase. ADP: Adenosine diphosphate. MFP: Membrane fusion protein found in the T1SS. OMP: Outer membrane protein.
2.1.1. Secretion Systems in Gram-Negative Bacteria
Type I Secretion System (T1SS):
The T1SS is prevalent in Gram-negative bacteria such as E. coli, Bordetella pertussis, and Vibrio cholerae. This system depends on the secretion of proteins by interaction with only three components. Two of them are found in the inner membrane (IM), an adenosine triphosphate (ATP) binding cassette transporter (ABC) and the membrane fusion protein (MFP), while the third component is found in the outer membrane (OM), the outer membrane protein (OMP) [18][19].
Each of these three components plays a role in the substrate’s secretion process. ABC transporters catalyze ATP to provide energy for the secretion process, contribute to substrate recognition, and interact with membrane fusion proteins (MFPs) in IM [20]. The MFP extends over the periphery of the plasma and connects the ABC transporter in IM with the OMF in OM. It contributes to substrate selection [2]. The role of the OMP is to form a long channel through the OM to the external environment to support substrate release [21].
Hemolysin A (HlyA) T1SS is the first protein for which secretion was discovered in bacteria. The protein is found in strains of E. coli that cause gastrointestinal and urinary tract infections [18]. HlyA destroys host cells by facilitating the invasion process, subsequently causing holes in the cell wall, leading to destruction. HlyA is transported by the T1SS, which consists of three HlyA transporters, HlyB, HlyD, and Tolc. The HlyB protein belongs to the ABC transporters family, is located at IM, and serves to catalyze APT to provide energy, while Tolc is located at OM and serves to form a long channel through OM to secrete HlyA into host cells. The HlyD protein serves as a connecting channel between HlyB and Tolc [22].
Type II Secretion System (T2SS):
T2SS is found in many Gram-negative bacteria, and it secretes folded proteins from the periplasm to the OM. The type II secretion system is an important attribute for non-pathogenic human strains such as Pseudomonas aeruginosa, E. coli, V. cholerae, Klebsiella species, Legionella pneumophila, and Yersinia enterocolitica. Proteins are secreted to the inner membrane via the general secretory pathway (Sec) or the twin-arginine translocation pathway (Tat), whereby they are secreted from the periplasm to the OM via the T2SS [23]. In addition, the Sec system must fold the proteins during transport before they enter the T2SS [2].
The Sec pathway is responsible for the transport of unfolded proteins to the outer environment, periplasm, or IM. It consists of three components that work together in the secretion of proteins: the motor protein, protein targeting, and SecYEG translocase. Many different proteins are transported through this membrane, some of which increase the virulence of bacteria. In addition, there are many pathogens that rely on the Sec pathway to transfer virulence factors from the periplasm. Proteins that use the Sec pathway to be transported to the surrounding membrane or to the external environment contain a (SecB) specific signaling sequence, while proteins that remain in the IM contain a specific signaling sequence (SRP) [2][23].
In contrast to the Sec pathway, the Tat pathway neglects the transport of mainly folded proteins, and this pathway was first characterized in the early 1990s in chloroplasts in plants [24]. In this pathway, proteins cannot be secreted if they are not folded [2]. In Gram-positive bacteria, the Tat pathway is used to transport folded proteins directly to the external environment, while in Gram-negative bacteria folded proteins are transported to the periplasm of the plasm and then transported to the external environment via T2SS [15].
Type III Secretion System (T3SS):
Host cells have some physiological processes that help protect them from infection, such as maintenance of the cytoskeleton of the cell, which includes maintaining the cell structure and the phagocyte; however, pathogens that contain type III secretion systems can disrupt all these processes in order to cause infection [25]. They can transfer proteins in a single step by passing through the IM and OM to excrete the proteins [2].
The structure of T3SS is complex, containing several units of about 20 bacterial proteins. There are proteins that transfer other proteins to the cytoplasm of the host cell, which are considered virulence factors, and these proteins are called translocators, while the translocated proteins are called effectors [25]. Many pathogens use methods to evade trapping by the host’s immune system. It was discovered that the type 3 mechanism of the secretion systemT3SS has an effect which enhances the ability of bacteria to escape from phagocytes [26].
Type V Secretion System (T5SS):
The T5SS is considered peculiar because it is very small compared to other types and the energy source that supplies the secretion process is unknown, since no chemicals such as ATP are found in the periplasm. It also does not contain a stable ion gradient across the OM, which is why it is called an autotransporter (AT). T5SS is present in the OM of bacteria, so it requires the transport of proteins through the Sec pathway first, and then it is transported from the OM to the external environment by T5SS [27][28]. The structure of T5SSs are different based on features of their field organization, so T5SS/ATs have been divided into a number of subtypes from Va to Ve, and recently there may also be Vf [29][30].
Type VI Secretion System (T6SS):
Gram-negative bacteria possess several secretion systems that assist them in the invasion of host cells, one of which is the T6SS. T6SS is one of the complex systems that transports the effector to the external environment through the bacterial membrane. The effector secreted by T6SS not only attacks eukaryotic cells, but also targets other bacteria in order to compete for survival and cause infection and disease [31][32]. Effector delivery methods are categorized, whereby the effectors can be classified as specialized effectors or cargo effectors. The specialized effector binds with the C-terminus of T6SS structure proteins such as Hcp, VgrG, or PAAR for translocation into target cells. It is believed that specialized effectors that fuse with Hcp-VgrG-PAA-associated effectors are released in a single lethal shot into other bacterial or eukaryotic cells [31][33][34]. Yersinia pseudotuberculosis has been studied to show a different feature in T6SS-4, in that it is able to transport zinc (Zn2+), which plays an important role in immune system resistance and anti-stress, contributing to the survival of bacteria in harmful environments. Zn2+ transport depends on the classic ZnuABC transporter with T6SS-4 and yezP, and the results showed that any disruption of these important transport elements leads to the bacteria losing their virulence against mice [35].
Type IX Secretion System (T9SS):
T9SS is responsible for the secretion of effector proteins such as proteases, adhesins, cellulases, chitin, and surface layer proteins. T9SS is found in species of periodontal pathogens such as Porphyromonas gingivalis that use T9SS to secrete up to 30 effector proteins. T9SS is the primary determinant of virulence of oral pathogenic bacteria species relevant to acute periodontal disease in humans and animals [36][37]. P. gingivalis and Tannerella forsythia are the most common pathogens of periodontal disease and chronic periodontitis. In T. forsythia, it secretes a KLIKK proteases which is a proteolytic enzyme by the T9SS. KLIKK proteases participate in the invasion of the host’s integumentary system in order to protect the bacteria, while in P. gingivalis, T9SS is used to secrete gingipains, which are proteolytic enzymes [38].
2.1.2. Secretion Systems in Both Gram-Negative Bacteria and Gram-Positive Bacteria
Type IV Secretion System (T4SS):
The T4SS is found in Gram-positive and Gram-negative bacteria, as well as in Archaea [39]. T4SS is characterized as being able to transport DNA along with proteins, whereby the DNA is transported without exposure to the outside of the cell. This system consists of two main components: the conjugation system and effector translocations. The conjugation system acts as a channel for the transfer of antibiotic resistance genes between bacteria, while effector translocations serve to transfer virulence factors to the host cell [15]. In Gram-negative bacteria, T4SS is divided into two types, T4SSA and T4SSB. In addition, Gram-negative and Gram-positive derived T4SSs were divided into eight classes based on detailed phylogenetic analysis [40]. It was discovered that the T4SS that is present in the Xanthomonadaceae family differs from the other T4SSs. The reason behind this is that the type IV secretion system present in Xanthomonas citri can secrete proteins that kill other Gram-negative bacteria in a contact-dependent manner [41]. T4SS was studied under a transmission electron microscope, where it was found that the basic molecular structure of the Dot/Icm secretion system type IV, consisting of at least five proteins encoded by DotC, DotD, DotF, DotG, and DotH, has a ring structure that is encoded by Legionella pneumophila, which is one of the intracellular opportunistic pathogens [42].
Type VII Secretion System (T7SS):
Mycobacteria and Corynebacteria species are resistant to the immune system and antibacterial drugs, and the reason for this is due to the presence of a thick waxy layer on the surface of the OM called the mycomembrane. There is a type VII bacterial secretion system which has effective mechanisms that help these bacteria secrete proteins through mycomembrane to the outside of the cell. The presence of the T7SS was first observed in M. tuberculosis. Recent research proved the presence of T7SS in non-mycomembrane bacteria such as S. aureus, Group B Streptococcus, Bacillus anthracis, and L. monocytogenes. The core T7SS structural components are encoded via gene clusters [2][43]. Type VII (T7SS) secretion systems in Group B Streptococcus contribute to the secretion of proteins that act as virulence factors, host toxicity, and killing of a number of bacteria in a few genera. Spencer and co-workers helped to understand the role of T7SS in the pathogenesis of GBS, as it is involved in a large part of the host’s cell interaction with Group B Streptococcus, as well as influencing the functions of the T7SS effector secreted by other Gram-positive bacteria [44]. Furthermore, the presence of T7SS in Staphylococcus aureus contributes to the resistance of antibacterial secreted by the host cells, as it has been shown that S. aureus lacking T7SS was killed by antimycotic fatty acids produced by the host cells [45].
2.2. Quorum Sensing (QS)
QS is a process of cell-to-cell communication through secretion of chemical signaling molecules that enable bacteria to recognize their surroundings [4]. QS helps bacteria regulate the population density of bacteria in their external environment by modifying their behavior through gene expression in response to chemical signals secreted by the bacteria. QS enables communication with other bacteria to live as multicellular organisms and collective interaction with host cells, and also helps meet the needs of given species for living in a particular niche [3][46][47]. The chemical signaling molecules called autoinducers are produced and released by bacteria, where different types of autoinducers are produced from different bacteria species as shown in Table 1. Bacteria then measure and respond to the accumulation of autoinducers in the environment. For example, when bacteria detect that the accumulation of a lower threshold concentration of these autoinducers has a stimulatory effect, they alter their gene expression to change their behavior accordingly [48].
Acute hepatopancreatic necrosis disease (AHPND), which is also called early mortality syndrome (EMS), appeared in Asia in 2009, which was the reason behind the destruction of the shrimp crop in East Asian countries and Mexico [49]. Vibrio spp. is the cause of AHPND, as it contains the pVA1 plasmid responsible for production of two toxins, PirAVP and PirBVP, that lead to symptoms of AHPND [50]. Bacteria precipitation may be affected by the environmental conditions around it. In the early log phase of the growth curve, the Vibrio strains showed an increase in the gene expression of PirAVP and PirBVP proteins, and this persisted into the log phase. It was discovered that the change in gene expression of PirAVP and PirBVP proteins was regulated by QS [51].
Table 1. Autoinducers molecules from different bacteria.
| Autoinducers | Microorganisms | Receptors | Phenotypes | References |
|---|---|---|---|---|
| Autoinducing peptides (AIPs) | Gram-positive bacteria | Response regulator | Genetic competence. | [52] |
| Autoinducer-2 (AI-2) | Many Gram-negative and Gram-positive bacteria | LuxP, LsrB, and dCACHE | Virulence, biofilm formation, and protease. | [53][54] |
| CAI-1 autoinducer | Vibrio | CqsS | Virulence, biofilm formation, and protease. | [53][55] |
| HAI-1 | Vibrio harveyi and Vibrio parahaemolyticus | LuxN | Biofilm formation, bioluminescence, TTS, and protease. | [56] |
| Acyl homoserine lactones (AHLs) | Gram-negative bacteria and commensals | LuxR | Elastase, biofilm formation, virulence factors, and exotoxins. | [57][58] |
| Competence-stimulating peptides (CSPs) | Streptococcus pneumoniae | ComD | Biofilm formation, and virulence. | [59] |
| comX-inducing peptide (XIP) | Streptococcus mutans | ComR | Antibiotic tolerance, genetic competence, and dormancy. | [60][61] |
| ComX | Bacillus subtilis | ComP | Protease and biofilm formation. | [62][63] |
2.2.1. QS in Gram-Negative Bacteria
QS is used by both Gram-negative bacteria and Gram-positive bacteria to communicate with each other and their environment, but there are differences in the mechanisms. First, the researchers will focus on quorum sensing in Gram-negative bacteria, and then later on quorum sensing in Gram-positive bacteria. There are some common features of quorum sensing in Gram-negative bacteria.
In Gram-negative organisms, autostimulants (AIs) are synthesized from S-adenosylmethionine (SAM) as molecules such as acyl-homoserine lactones (AHLs), which are the most common class of AIs. They consist of an acyl-homoserine lactone ring and an acyl-carbon chain, the length of which affects the stability of the AHLs. LuxI family enzymes are the main producers of AHLs. They function as AHL synthases and catalyze the acylation of SAM by the acyl carrier protein (acyl-ACP). In this reaction, SAM provides the amino group and the acyl-ACP provides the acyl group for the synthesis of AHLs, which are released to the outside via the cell membrane and act as co-signaling molecules [46][64][65][66].
There are specific receptors in the cytoplasm and IM that associate with autoinducers; these receptors associated with AHLs aid gene expression in QS. For example, LuxR-type receptors in the cytoplasm are transcription factors. LuxR binds with AHLs to form a complex that binds to DNA to regulate bacterial gene expression. In P. aeruginosa, the LuxR/LuxI-type system contributes to communication with other cells [67][68].
Furthermore, receptors in the IM and cytoplasm regulate hundreds of gene expressions through QS. Receptors can regulate genes that directly influence the formation of virulence, biofilm, and the biological and physiological processes of bacteria. QS receptor molecules regulate gene expression through a mechanism called autoinduction. It is a mechanism that enhances gene expression in all bacterial populations by increasing the synthesis of autoinducers, resulting in an increased population at the end of the response [68].
2.2.2. Quorum Sensing in Gram-Positive Bacteria
QS in Gram-positive bacteria uses short peptides as autoinducers to determine population density as shown in Figure 2 [69]. These autoinducer peptides (AIP) are synthesized by the ribosome and then modified to be active. It is released into the external environment with the help of the ABC transporter located in the IM [70]. Autoinducers accumulate in the external environment until a threshold concentration is reached, and this is recognized by a specific protein, histidine kinase. Then gene expression is preceded in the bacterial population in response to the concentration of autoinducers. The higher the concentration of autoinducers in the external environment, the greater the bacterial population density [71].
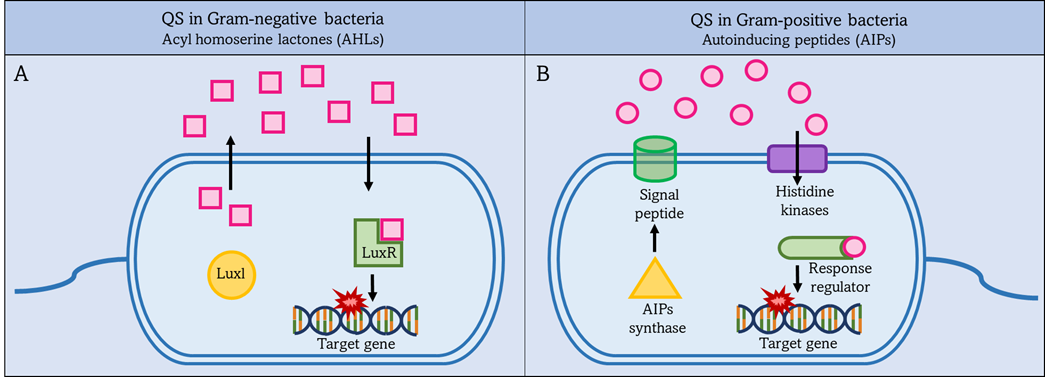
Figure 2. QS systems in Gram-negative and Gram-positive bacteria. Left figure (A): Gram-negative bacteria secrete acyl-homoserine lactones (AHLs) as autoinducers, which are synthesized by Luxl and then pass through the bacterial membrane into the external environment. Once the AHLs reach a threshold level, they activate intracellular LuxR to activate target gene expression. Right figure (B): Gram-positive bacteria secrete autoinduction peptides (AIPs), which are synthesized by AIPs synthase and then pass through the bacterial membrane into the external environment. Once the AIPs reach a threshold level. This is detected by a specific protein, histidine kinases, and activates the intracellular regulator. The response regulator leads to increased expression of the target gene.
There is a family of transcriptional regulators associated with peptides to help coordinate gene expression. This is the RRNPP family, which consists of Rap phosphatases, NprR sensor, PlcR, PrgX, and finally Rgg, which was added later. The RRNPP family is found in Bacillus, Streptococcus, and Enterococcus [71][72]. It was determined that there is a relationship between QS and the transformation of commensals into pathogens in S. pneumoniae. Streptococci species Rgg/SHP is widely spread among their species, as it was discovered that the S. pneumoniae D39 Rgg0939/SHP system regulates transcription of one gene group, and it was established that QS in Streptococci species is responsible for regulating exopolysaccharide synthesis from a distinct site [73].
2.2.3. The Role of Quorum Sensing in Biofilm Formation
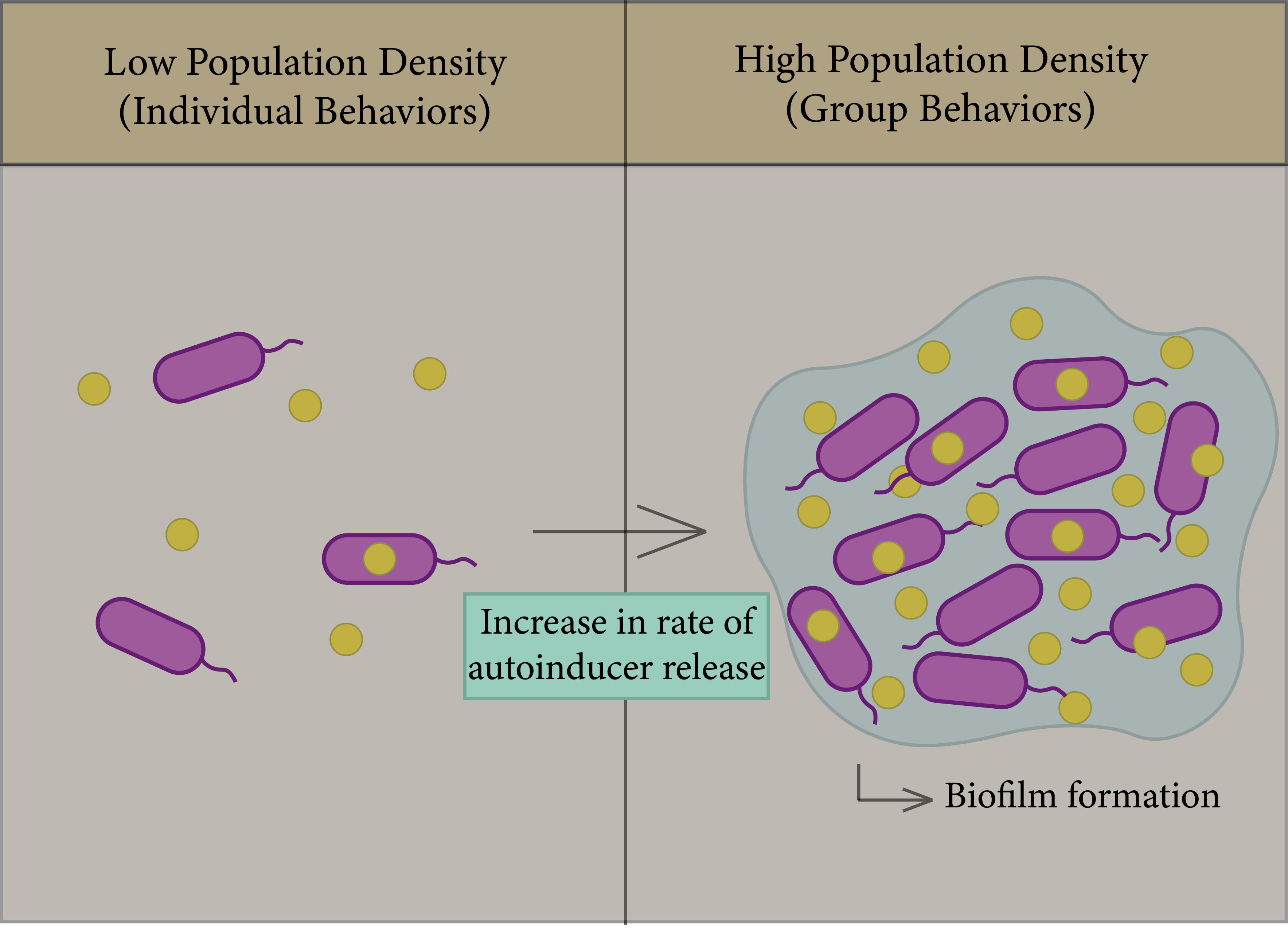
In bacteria, biofilms are one of the most important factors that lead to antibiotic resistance and a reason for persistence of infection [74]; the general principle of biofilm formation is illustrated in Figure 3. Biofilms consist of a group of the same or different types of bacteria that adhere to each other and to the surface, whereby they assemble together by polymeric matrices consisting of secreted proteins, sugars, lipids, and extracellular DNA [75]. QS helps in increasing the rate of biofilm formation, in addition to virulence factors. Bacteria can trap secreted autoinducers from the quorum sensor in biofilm gene expression in order to enhance bacterial survival in the colonizing environment [74][76][77].
Figure 3. General principle of the QS system. Left figure: Each species begins to secrete autoinducers into the external environment, and the population density increases. Right figure: Increased rate of autoinducer release to form biofilm due to gene expression.
Biofilms are formed through four steps: binding, cell-to-cell adhesion, maturation, and dispersal [74]. Once biofilms are formed, they are accompanied by their extirpation, as they have the ability to resist antimicrobials, as the formation of cell membranes is one of the mechanisms of bacterial adaptation to their environment. The study of Streptococcus gordonii, which demonstrated the role of QS systems in the formation of cell membranes, found a defective mutation in biofilms which have a transposon insertion in the comD gene, which is responsible for encoding the sensor protein (histidine kinase) in Gram-positive bacterial QS. As such, the formation of biofilm is highly dependent on communication between cells via QS [77][78].
References
- Ralf Koebnik; Kaspar P. Locher; Patrick Van Gelder; Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Molecular Microbiology 2000, 37, 239-253, 10.1046/j.1365-2958.2000.01983.x.
- Erin R. Green; Joan Mecsas; Bacterial Secretion Systems: An Overview. Microbiology Spectrum 2016, 4, 213-239, 10.1128/microbiolspec.vmbf-0012-2015.
- Melissa B. Miller; Bonnie L. Bassler; Quorum Sensing in Bacteria. Annual Review of Microbiology 2001, 55, 165-199, 10.1146/annurev.micro.55.1.165.
- Bonnie L. Bassler; Richard Losick; Bacterially Speaking. Cell 2006, 125, 237-246, 10.1016/j.cell.2006.04.001.
- Julian G. Hurdle; Alex O'Neill; Ian Chopra; Richard Lee; Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nature Reviews Genetics 2010, 9, 62-75, 10.1038/nrmicro2474.
- Ido M. Herzog; Micha Fridman; Design and synthesis of membrane-targeting antibiotics: from peptides- to aminosugar-based antimicrobial cationic amphiphiles. MedChemComm 2014, 5, 1014-1026, 10.1039/c4md00012a.
- Rimantas Daugelavičius; Elena Bakiene˙; Dennis H. Bamford; Stages of Polymyxin B Interaction with the Escherichia coli Cell Envelope. Antimicrobial Agents and Chemotherapy 2000, 44, 2969-2978, 10.1128/aac.44.11.2969-2978.2000.
- Chalermrat Bunchorntavakul; Naichaya Chamroonkul; Disaya Chavalitdhamrong; Bacterial infections in cirrhosis: A critical review and practical guidance. World Journal of Hepatology 2016, 8, 307-21, 10.4254/wjh.v8.i6.307.
- Laura A. Dever; Terence S. Dermody; Mechanisms of Bacterial Resistance to Antibiotics. Archives of Internal Medicine 1991, 151, 886-895, 10.1001/archinte.1991.00400050040010.
- Carole Ayoub Moubareck; Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181, 10.3390/membranes10080181.
- Peterson, J.W. Bacterial Pathogenesis. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2.
- J W Wilson; M J Schurr; C L Leblanc; R Ramamurthy; K L Buchanan; C A Nickerson; Mechanisms of bacterial pathogenicity. Postgraduate Medical Journal 2002, 78, 216-224, 10.1136/pmj.78.918.216.
- Norrby, S.R. Chapter 54—Urinary Tract Infections. In Antibiotic and Chemotherapy, 9th ed.; Finch, R.G., Greenwood, D., Norrby, S.R., Whitley, R.J., Eds.; W.B. Saunders: London, UK, 2010; pp. 694–701. ISBN 978-0-7020-4064-1
- S. Doron; S.L. Gorbach; Bacterial Infections: Overview. null 2008, ,, 273-282, 10.1016/b978-012373960-5.00596-7.
- Rocio Trastoy Pena; Lucía Blasco; Antón Ambroa; Bertha González-Pedrajo; Laura Fernández-García; Maria López; Ines Bleriot; German Bou; Rodolfo García-Contreras; Thomas Keith Wood; et al.Maria Tomás Relationship Between Quorum Sensing and Secretion Systems. Frontiers in Microbiology 2019, 10, 1100, 10.3389/fmicb.2019.01100.
- Rhea G. Abisado; Saida Benomar; Jennifer R. Klaus; Ajai A. Dandekar; Josephine R. Chandler; Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17, 10.1128/mbio.02331-17.
- Sofie Depluverez; Simon Devos; Bart Devreese; The Role of Bacterial Secretion Systems in the Virulence of Gram-Negative Airway Pathogens Associated with Cystic Fibrosis. Frontiers in Microbiology 2016, 7, 1336, 10.3389/fmicb.2016.01336.
- Sabrina Thomas; I. Barry Holland; Lutz Schmitt; The Type 1 secretion pathway — The hemolysin system and beyond. Biochimica et Biophysica Acta 2013, 1843, 1629-1641, 10.1016/j.bbamcr.2013.09.017.
- Jacob L.W. Morgan; Justin F. Acheson; Jochen Zimmer; Structure of a Type-1 Secretion System ABC Transporter. Structure 2017, 25, 522-529, 10.1016/j.str.2017.01.010.
- Kerstin Kanonenberg; Christian K.W. Schwarz; Lutz Schmitt; Type I secretion systems – a story of appendices. Research in Microbiology 2013, 164, 596-604, 10.1016/j.resmic.2013.03.011.
- P. Delepelaire; Type I secretion in gram-negative bacteria. Biochimica et Biophysica Acta 2004, 1694, 149-161, 10.1016/j.bbamcr.2004.05.001.
- Minho Lee; So-Young Jun; Bo-Young Yoon; Saemee Song; Kangseok Lee; Nam-Chul Ha; Membrane Fusion Proteins of Type I Secretion System and Tripartite Efflux Pumps Share a Binding Motif for TolC in Gram-Negative Bacteria. PLOS ONE 2012, 7, e40460, 10.1371/journal.pone.0040460.
- Konstantin V. Korotkov; Maria Sandkvist; Wim G. J. Hol; The type II secretion system: biogenesis, molecular architecture and mechanism. Nature Reviews Genetics 2012, 10, 336-351, 10.1038/nrmicro2762.
- Colin Robinson; Albert Bolhuis; Tat-dependent protein targeting in prokaryotes and chloroplasts. Physica D: Nonlinear Phenomena 2004, 1694, 135-147, 10.1016/s0167-4889(04)00086-2.
- Bryan Coburn; Inna Sekirov; B. Brett Finlay; Type III Secretion Systems and Disease. Clinical Microbiology Reviews 2007, 20, 535-549, 10.1128/cmr.00013-07.
- Juan Du; Analise Z. Reeves; Jessica A. Klein; Donna J. Twedt; Leigh A. Knodler; Cammie F. Lesser; The type III secretion system apparatus determines the intracellular niche of bacterial pathogens. Proceedings of the National Academy of Sciences 2016, 113, 4794-4799, 10.1073/pnas.1520699113.
- Ina Meuskens; Athanasios Saragliadis; Jack C. Leo; Dirk Linke; Type V Secretion Systems: An Overview of Passenger Domain Functions. Frontiers in Microbiology 2019, 10, 1163, 10.3389/fmicb.2019.01163.
- Jack Leo; Iwan Grin; Dirk Linke; Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philosophical Transactions of the Royal Society B: Biological Sciences 2012, 367, 1088-1101, 10.1098/rstb.2011.0208.
- Jan Grijpstra; Jesús Arenas; Lucy Rutten; Jan Tommassen; Autotransporter secretion: varying on a theme. Research in Microbiology 2013, 164, 562-582, 10.1016/j.resmic.2013.03.010.
- Ian R. Henderson; Fernando Navarro-Garcia; Mickaël Desvaux; Rachel C. Fernandez; Dlawer Ala'Aldeen; Type V Protein Secretion Pathway: the Autotransporter Story. Microbiology and Molecular Biology Reviews 2004, 68, 692-744, 10.1128/mmbr.68.4.692-744.2004.
- Yun-Wei Lien; Erh-Min Lai; Type VI Secretion Effectors: Methodologies and Biology. Frontiers in Cellular and Infection Microbiology 2017, 7, 254-254, 10.3389/fcimb.2017.00254.
- Brian T. Ho; Tao G. Dong; John J. Mekalanos; A View to a Kill: The Bacterial Type VI Secretion System. Cell Host & Microbe 2013, 15, 9-21, 10.1016/j.chom.2013.11.008.
- Fernando Navarro-Garcia; Fernando Ruiz-Perez; Ángel Cataldi; Mariano Larzabal; Type VI Secretion System in Pathogenic Escherichia coli: Structure, Role in Virulence, and Acquisition. Frontiers in Microbiology 2019, 10, 1965, 10.3389/fmicb.2019.01965.
- Francesca R. Cianfanelli; Laura Monlezun; Sarah J. Coulthurst; Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends in Microbiology 2015, 24, 51-62, 10.1016/j.tim.2015.10.005.
- Tietao Wang; Meiru Si; Yunhong Song; Wenhan Zhu; Fen Gao; Yao Wang; Lei Zhang; Weipeng Zhang; Gehong Wei; Zhao-Qing Luo; et al.Xihui Shen Type VI Secretion System Transports Zn2+ to Combat Multiple Stresses and Host Immunity. PLOS Pathogens 2015, 11, e1005020, 10.1371/journal.ppat.1005020.
- Dhana G. Gorasia; Paul D. Veith; Eric C. Reynolds; The Type IX Secretion System: Advances in Structure, Function and Organisation. Microorganisms 2020, 8, 1173, 10.3390/microorganisms8081173.
- Frederic Lauber; Justin Deme; Susan M. Lea; Ben C. Berks; Type 9 secretion system structures reveal a new protein transport mechanism. Nature Cell Biology 2018, 564, 77-82, 10.1038/s41586-018-0693-y.
- Lahari Koneru; Type IX secretion system : characterization of an effector protein and an insight into the role of c-terminal domain dimeration in outer membrane translocation.. Type IX secretion system : characterization of an effector protein and an insight into the role of c-terminal domain dimeration in outer membrane translocation. 2018, -, 2854, 10.18297/etd/2854.
- Eric Cascales; Peter J. Christie; The versatile bacterial type IV secretion systems. Nature Reviews Genetics 2003, 1, 137-149, 10.1038/nrmicro753.
- Germán Gustavo Sgro; Gabriel U. Oka; Diorge Souza; William Cenens; Ethel Bayer-Santos; Bruno Y. Matsuyama; Natalia F. Bueno; Thiago Rodrigo Dos Santos; Cristina Elisa Alvarez-Martinez; Roberto K. Salinas; et al.Chuck S. Farah Bacteria-Killing Type IV Secretion Systems. Frontiers in Microbiology 2019, 10, 1078, 10.3389/fmicb.2019.01078.
- Diorge Souza; Gabriel U. Oka; Cristina Elisa Alvarez-Martinez; Alexandre Wilson Bisson Filho; German Dunger; Lise Hobeika; Nayara S. Cavalcante; Marcos C. Alegria; Leandro Barbosa; Roberto Kopke Salinas; et al.Cristiane R. GuzzoChuck Shaker Farah Bacterial killing via a type IV secretion system. Nature Communications 2015, 6, 6453, 10.1038/ncomms7453.
- Tomoko Kubori; Masafumi Koike; Xuan Thanh Bui; Saori Higaki; Shin-Ichi Aizawa; Hiroki Nagai; Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proceedings of the National Academy of Sciences 2014, 111, 11804-11809, 10.1073/pnas.1404506111.
- Edith N.G. Houben; Konstantin Korotkov; Wilbert Bitter; Take five — Type VII secretion systems of Mycobacteria. Biochimica et Biophysica Acta 2014, 1843, 1707-1716, 10.1016/j.bbamcr.2013.11.003.
- Brady L. Spencer; Uday Tak; Jéssica C. Mendonça; Prescilla E. Nagao; Michael Niederweis; Kelly S. Doran; A type VII secretion system in Group B Streptococcus mediates cytotoxicity and virulence. PLOS Pathogens 2021, 17, e1010121, 10.1371/journal.ppat.1010121.
- Arnaud Kengmo Tchoupa; Kate E. Watkins; Rebekah A. Jones; Agnès Kuroki; Mohammad Tauqeer Alam; Sebastien Perrier; Yin Chen; Meera Unnikrishnan; The type VII secretion system protects Staphylococcus aureus against antimicrobial host fatty acids. Scientific Reports 2020, 10, 1-16, 10.1038/s41598-020-71653-z.
- Liang Wu; Yubin Luo; Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Frontiers in Microbiology 2021, 12, e611413, 10.3389/fmicb.2021.611413.
- Nicola C. Reading; Vanessa Sperandio; Quorum sensing: the many languages of bacteria. FEMS Microbiology Letters 2006, 254, 1-11, 10.1111/j.1574-6968.2005.00001.x.
- Christopher M. Waters; Bonnie L. Bassler; QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annual Review of Cell and Developmental Biology 2005, 21, 319-346, 10.1146/annurev.cellbio.21.012704.131001.
- L Nunan; D Lightner; C Pantoja; S Gomez-Jimenez; Detection of acute hepatopancreatic necrosis disease (AHPND) in Mexico. Diseases of Aquatic Organisms 2014, 111, 81-86, 10.3354/dao02776.
- Phuong Thi Ngoc Tran; Vikash Kumar; Peter Bossier; Do acute hepatopancreatic necrosis disease-causing PirABVP toxins aggravate vibriosis?. Emerging Microbes & Infections 2020, 9, 1919-1932, 10.1080/22221751.2020.1811778.
- Shin-Jen Lin; Jiun-Yan Huang; Phuoc-Thien Le; Chung-Te Lee; Che-Chang Chang; Yi-Yuan Yang; Emily Chia-Yu Su; Chu-Fang Lo; Hao-Ching Wang; Expression of the AHPND Toxins PirAvp and PirBvp Is Regulated by Components of the Vibrio parahaemolyticus Quorum Sensing (QS) System. International Journal of Molecular Sciences 2022, 23, 2889, 10.3390/ijms23052889.
- Lantian Zhou; Yue Zhang; Yongze Ge; Xuan Zhu; Jianyi Pan; Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Frontiers in Microbiology 2020, 11, 589640, 10.3389/fmicb.2020.589640.
- Brian Hammer; Bonnie L. Bassler; Quorum sensing controls biofilm formation in Vibrio cholerae. Molecular Microbiology 2003, 50, 101-104, 10.1046/j.1365-2958.2003.03688.x.
- Lei Zhang; Shuyu Li; Xiaozhen Liu; Zhuo Wang; Mei Jiang; Ruiying Wang; LaiGong Xie; Qinmeng Liu; Xiaorong Xie; Daohan Shang; et al.Mengyun LiZhiyan WeiYao WangChengpeng FanZhao-Qing LuoXihui Shen Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nature Communications 2020, 11, 1-13, 10.1038/s41467-020-19243-5.
- Wai‐Leung Ng; Lark J. Perez; Yunzhou Wei; Christina Kraml; Martin F. Semmelhack; Bonnie L. Bassler; Signal production and detection specificity inVibrioCqsA/CqsS quorum‐sensing systems. Molecular Microbiology 2011, 79, 1407-1417, 10.1111/j.1365-2958.2011.07548.x.
- Christopher M. Waters; Bonnie L. Bassler; The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes & Development 2006, 20, 2754-2767, 10.1101/gad.1466506.
- Garance Coquant; Jean-Pierre Grill; Philippe Seksik; Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Frontiers in Immunology 2020, 11, 1827, 10.3389/fimmu.2020.01827.
- Lisheng Liao; Amy L. Schaefer; Bruna G. Coutinho; Pamela J. B. Brown; E. Peter Greenberg; An aryl-homoserine lactone quorum-sensing signal produced by a dimorphic prosthecate bacterium. Proceedings of the National Academy of Sciences 2018, 115, 7587-7592, 10.1073/pnas.1808351115.
- Yifang Yang; Jingjun Lin; Anthony Harrington; Gabriel Cornilescu; Gee W. Lau; Yftah Tal-Gan; Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity in Streptococcus pneumoniae. Proceedings of the National Academy of Sciences 2020, 117, 1689-1699, 10.1073/pnas.1915812117.
- Vincent Eleung; Delphine Edufour; Céline M. Lévesque; Death and survival in Streptococcus mutans: differing outcomes of a quorum-sensing signaling peptide. Frontiers in Microbiology 2015, 6, 1176, 10.3389/fmicb.2015.01176.
- Chowdhury Raihan Bikash; Yftah Tal-Gan; Structure Activity Relationship Study of the XIP Quorum Sensing Pheromone in Streptococcus mutans Reveal Inhibitors of the Competence Regulon. ACS Chemical Biology 2020, 15, 2833-2841, 10.1021/acschembio.0c00650.
- Margarita Kalamara; Mihael Spacapan; Ines Mandic-Mulec; Nicola R. Stanley-Wall; Social behaviours byBacillus subtilis: quorum sensing, kin discrimination and beyond. Molecular Microbiology 2018, 110, 863-878, 10.1111/mmi.14127.
- Mihael Špacapan; Tjaša Danevčič; Polonca Štefanic; Michael Porter; Nicola R. Stanley-Wall; Ines Mandic-Mulec; The ComX Quorum Sensing Peptide of Bacillus subtilis Affects Biofilm Formation Negatively and Sporulation Positively. Microorganisms 2020, 8, 1131, 10.3390/microorganisms8081131.
- Kai Papenfort; Bonnie L. Bassler; Quorum sensing signal–response systems in Gram-negative bacteria. Nature Reviews Genetics 2016, 14, 576-588, 10.1038/nrmicro.2016.89.
- Youai Hao; Stephen C. Winans; Bernard R. Glick; Trevor C. Charles; Identification and characterization of new LuxR/LuxI-type quorum sensing systems from metagenomic libraries. Environmental Microbiology 2009, 12, 105-117, 10.1111/j.1462-2920.2009.02049.x.
- Xue Li; Gongliang Zhang; Yaolei Zhu; Jingran Bi; Hongshun Hao; Hongman Hou; Effect of the luxI/R gene on AHL-signaling molecules and QS regulatory mechanism in Hafnia alvei H4. AMB Express 2019, 9, 1-11, 10.1186/s13568-019-0917-z.
- Ashraf Kariminik; Majid Baseri-Salehi; Babak Kheirkhah; Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article. Immunology Letters 2017, 190, 1-6, 10.1016/j.imlet.2017.07.002.
- Steven T. Rutherford; Bonnie L. Bassler; Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspectives in Medicine 2012, 2, a012427-a012427, 10.1101/cshperspect.a012427.
- Frederick Verbeke; Severine De Craemer; Nathan Debunne; Yorick Janssens; Evelien Wynendaele; Christophe Van de Wiele; Bart De Spiegeleer; Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo. Frontiers in Neuroscience 2017, 11, 183, 10.3389/fnins.2017.00183.
- Mark H.J. Sturme; Michiel Kleerebezem; Jiro Nakayama; Antoon D.L. Akkermans; Elaine E. Vaughan; Willem M. De Vos; Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie van Leeuwenhoek 2002, 81, 233-243, 10.1023/a:1020522919555.
- Véronique Monnet; Rozenn Gardan; Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide ’. Molecular Microbiology 2015, 97, 181-184, 10.1111/mmi.13060.
- David Perez-Pascual; Véronique Monnet; Rozenn Gardan; Bacterial Cell–Cell Communication in the Host via RRNPP Peptide-Binding Regulators. Frontiers in Microbiology 2016, 7, 706, 10.3389/fmicb.2016.00706.
- Roger Junges; Gabriela Salvadori; Sudhanshu Shekhar; Heidi A. Åmdal; Jimstan N. Periselneris; Tsute Chen; Jeremy S. Brown; Fernanda C. Petersen; A Quorum-Sensing System That Regulates Streptococcus pneumoniae Biofilm Formation and Surface Polysaccharide Production. mSphere 2017, 2, e00324-17, 10.1128/msphere.00324-17.
- Veronica Georgiana Preda; Oana Săndulescu; Communication is the key: biofilms, quorum sensing, formation and prevention. Discoveries 2019, 7, e10, 10.15190/d.2019.13.
- Yannick Dn Tremblay; Cynthia Lévesque; Ruud Pam Segers; Mario Jacques; Method to grow Actinobacillus pleuropneumoniaebiofilm on a biotic surface. BMC Veterinary Research 2013, 9, 213-213, 10.1186/1746-6148-9-213.
- Tsiry Rasamiravaka; Quentin Labtani; Pierre Duez; Mondher El Jaziri; The Formation of Biofilms byPseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Research International 2015, 2015, 1-17, 10.1155/2015/759348.
- Nira Rabin; Yue Zheng; Clement Opoku-Temeng; Yixuan Du; Eric Bonsu; Herman O Sintim; Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Medicinal Chemistry 2015, 7, 493-512, 10.4155/fmc.15.6.
- D L Chopp; M J Kirisits; B Moran; M R Parsek; A mathematical model of quorum sensing in a growing bacterial biofilm. Journal of Industrial Microbiology and Biotechnology 2002, 29, 339-346, 10.1038/sj/jim/7000316.