Despite tremendous advancements in technologies and resources, drug discovery still remains a tedious and expensive process. Though most cells are cultured using 2D monolayer cultures, due to lack of specificity, biochemical incompatibility, and cell-to-cell/matrix communications, they often lag behind in the race of modern drug discovery. There exists compelling evidence that 3D cell culture models are quite promising and advantageous in mimicking in vivo conditions. It is anticipated that these 3D cell culture methods will bridge the translation of data from 2D cell culture to animal models. Although 3D technologies have been adopted widely these days, they still have certain challenges associated with them, such as the maintenance of a micro-tissue environment similar to in vivo models and a lack of reproducibility. However, newer 3D cell culture models are able to bypass these issues to a maximum extent.
1. Introduction
Drug discovery and development is a lengthy and expensive process due to the high attrition rate in the clinical success of therapeutic agents
[1]. To improve drug discovery success rates, newer technologies with higher precision are required. Two traditional and promising approaches in drug discovery include biochemical assays and cell-based assays
[2]. Biochemical assays are straightforward and consistent methods to screen out compounds with an expected therapeutic potential towards a target enzyme or receptor
[3]. On the other hand, cell-based assays are more complicated and utilised for functional aspects in a cellular framework
[4]. Traditionally, cell-based assays were performed in two-dimensional (2D) monolayer cells cultured on various types of planar substrates
[5]. The 2D cultures were predominantly used for cell-based high throughput screening to discover drug-like molecules. Currently, these 2D cell models are reliable and very effective approaches for predicting responses of various drugs in vivo as well as for understanding vital molecular and underlying cellular mechanisms
[6]. Also, these models have been successfully employed in studying disease pathologies and biomarker discovery
[7]. Although, the monolayer models have had vast utilisation in the past, they are still not able to reiterate major in vivo facets, leading to their limited utilization in the modern drug discovery process. Beyond this, the 2D models also have other limitations such as lack of tissue-specificity, mechanical issues, biochemical disturbances, and cell-to-cell/cell-to-matrix-incompatibilities
[8][9]. All these issues reveal them to be weaker models to envisage drug efficacy for some specific diseases like cancer.
The newer three-dimensional (3D) cell culture techniques have been widely explored in the past decade in drug development, which has led to improved precision and a reduced failure rate of drugs in clinical phases. The accomplishment of 3D-culture models in early drug discovery has been widely adopted nowadays by the pharmaceutical research and development sectors
[10][11][12]. It is well established now that 3D-culture systems mimic the tissue factors and are the best representatives of the in vivo cellular phenomena in comparison to 2D models
[13][14]. One of the greatest advantages of 3D models is that, together with stem cells or primary cell models, they are able to predict the efficacy as well as toxicity of therapeutic candidates in humans before drugs enter clinical trials
[15]. Hence, these are contributing greatly to reducing the attrition rate of drug discovery and development processes. During 3D-culture experiments, as the cell culture mimics the in vivo cellular atmosphere, more efficient observations related to cell-to-cell interactions, tumor properties, metabolomics, stem cell research, and pathophysiology of many other diseases can be studied
[16].
2. 3D Cell Culture Technologies
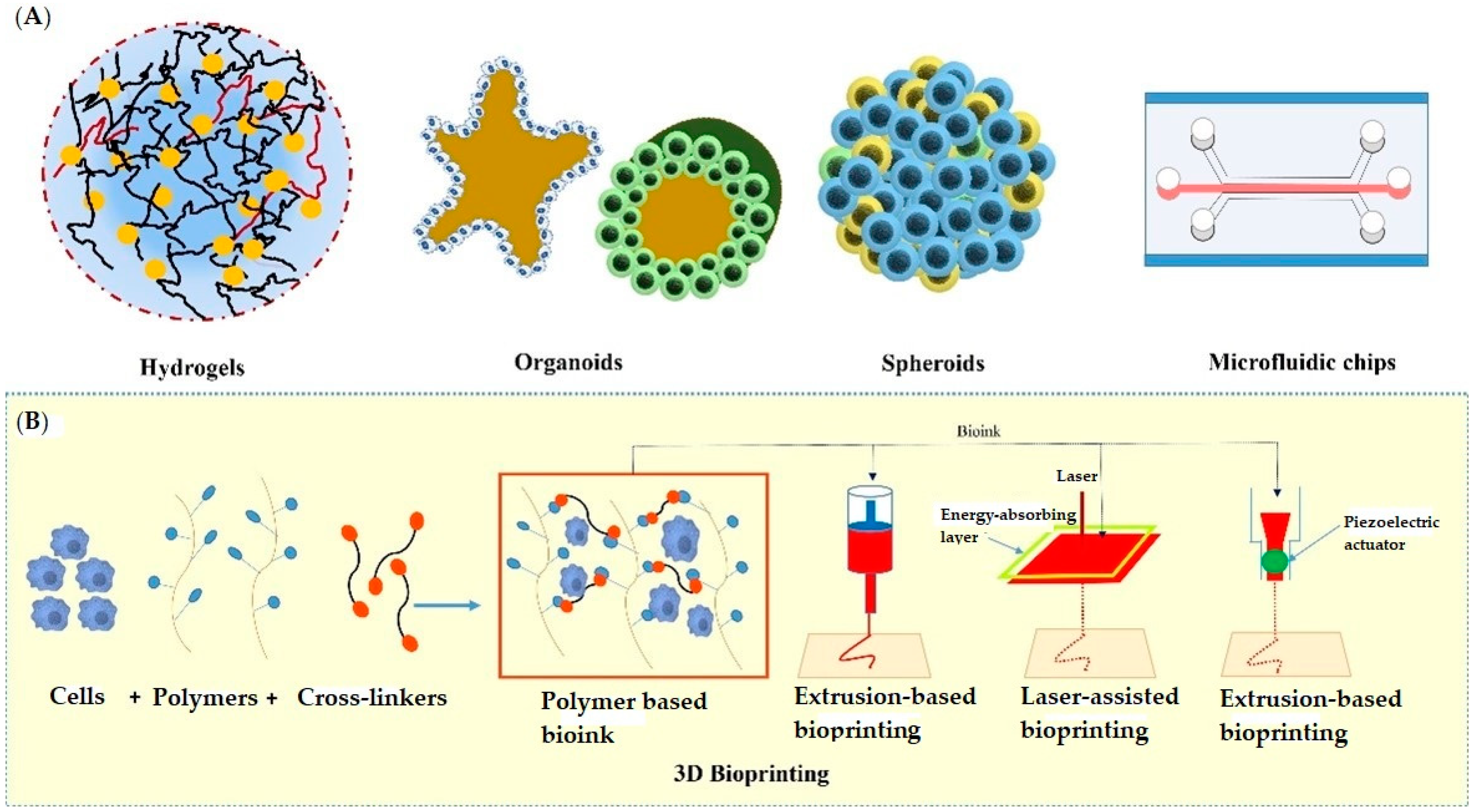
The past few years have evidenced success stories of development in 3D-culture models that imitate in vivo physiology. These technologies have been widely adopted in cell biology, tissue engineering, as well as in several clinical investigations. The significant 3D approaches include multicellular spheroids, organoids, scaffold approaches, microfluidics/lab on chip techniques, 3D bioprinting, hydrogels, anchorage approaches, hanging drop microplates, magnetic levitation, etc. All these approaches have been utilized by several research groups in drug discovery and development (discussed below). Figure 1 depicts various 3D cell culturing methods.
Figure 1. Various approaches used in 3D cell culturing; (A) showing hydrogels, organoids, spheroids, and microfluidic chip, (B) showing the extrusion-based, laser-assisted, and inkjet-based methods used in 3D Bioprinting.
2.1. Hydrogels
Due to their greater veracity, hydrogels have achieved a popular place in ex vivo tissue-like structure cultivation of cells, thereby becoming ideal candidates for 3D culture of tissues due to their similarities with the biological properties of extracellular matrices (ECM)
[17]. In general, hydrogels are made up of crosslinked polymers that can absorb large amounts of water (about 95% of their weight) along with other solutes within their swollen matrices, thereby allowing sustained delivery of absorbed solutes. These polymers exhibit desirable properties such as diverse mechanisms, different mesh sizes, puffiness, and deprivations that provide a significant advantage in 3D cell culture models
[14]. They can exist as a polymer molecular network due to the presence of intermolecular crosslinks or as fibrillar hydrogels formed via interfibrillar crosslinks
[18][19]. The hydrogels can be classified into various categories based on their structural properties, degradability, molecular charge, responsiveness to external stimuli, and source of production
[20].
The most common biological source-derived hydrogels used as extracellular matrices proteins are collagens, Matrigel, and alginate. the source of these hydrogels is biological, they are more compatible with 3D cell culturing
[21]. The complex nature of these hydrogels makes them non-tuneable, which is disadvantageous in the design of 3D models with required features. To overcome this issue, a number of synthetically produced hydrogels have been reported by various research groups with tuneable material properties as required in 3D cell culturing.
Sawhney et al. reported poly(ethylene glycol)-based photopolymerized bioerodible macromolecules as hydrogels with successful application in 3D cell culturing
[22]. These hydrogels were found to be effective at maintaining encapsulated cell viability, reproducibility, tuneable mechanical properties, and low cost production
[23]. Zustiak and co. give PEG derived hydrolytically degradable hydrogels with tuneable, degradable and mechanical properties
[24]. Findings from the study suggested that the properties of hydrogels can be controlled by altering polymer density, molecular weight, and the distance between the ester and thiol group in the cross-linker at the same time as keeping hydrogel repeat units and functional groups constant to maintain the cross-linking and degradation conditions compatible for protein and cell encapsulation. Martens et al. reported acrylated poly(vinyl alcohol) macromolecules as hydrogels synthesized via photopolymerization crosslinking
[25] and revealed that the choice of crosslinking method greatly influences the polymer network structure in several aspects. Horak and his group successfully employed these hydrogels in a mouse embryonic stem cell model
[26]. Notably, polyacrylamide hydrogels matrices posesses easily quantifiable elasticity, which can be modified by adjusting the relative concentrations of the monomer (i.e., acrylamide) and cross-linker (bis-acrylamide). Tse et al. have given a photoinitiated polymerization method for polyacrylamide derived hydrogels with tuneable mechanical properties via varying concentrations of acrylamides due to fabrication
[27]. Some other researchers also synthesized hydrogels include poly(ethylene oxide) (PEO), poly(methacrylic acid) (PMMA)
[28], poly propylene furmarate-co-ethylene glycol (P(PF-co-EG))
[29][30], poly(acrylamide) (PAAm)
[27], poly N-isopropylacrylamide (PNIPAAm)
[31], etc.
[32]. The low-cost production, consistency, and tuneable properties have made them the centre of attraction in 3D cell culture. In contrast, these synthetic hydrogels are less biocompatible as compared to hydrogels obtained from natural sources due to a lack of endogenous biological moieties
[33]. The biocompatibility of synthetic hydrogels can also be enhanced via choosing a compatible starting material, but it can increase the cost depending on the synthetic procedure.
2.2. Spheroids
Spheroids are cell aggregates that can self-assemble in an environment that usually does not allow adhesion to a smooth surface
[34]. Spheroid cultures were first developed around 1970 as multicellular cultures to reiterate the phenotype of human cancer cells and their retort to radiation therapy
[35]. After that, spheroid cultures have been utilized for a wide variety of cells, such as stem, hepatic, and neuronal cells. Contrary to the monolayer cultures, 3D spheroids exhibit heterogeneous cell colonies, i.e., cells at proliferating, quiescent, hypoxic, apoptotic, and necrotic stages. The outer layers that are highly exposed to the medium comprise viable and proliferating cells, whereas the core cells tend to be in a hypoxic or quiescent state as they receive less oxygen, nutrients, and other essential compounds from the medium
[36]. Also, these have a definite geometry and well-defined cell-to-cell and cell-to-matrix communications. Various membrane (integrins) and extracellular matrix proteins are responsible for the formation of spheroids. Spheroid construction involves aggregation of dispersed cells resulting from long-chain extracellular fibres, allowing binding of surface integrins, leading to upregulation of cadherin expression
[37][38]. Further, this cadherin gets deposited on the cell membrane surface, which is responsible for haemophilic cadherin-cadherin interactions, forming tight connections between adjacent cells, resulting in spheroid formation. Finally, the integrins are involved in the activation of focal adhesion kinase (FAK), the overexpression of which has been linked to tumor growth
[39][40]. The spheroids have been recently utilized in various drug discovery processes against a variety of diseases, especially in oncology research. Also, the 3D-spheroids can be employed to study the metabolic processes in both intra-cellular and extra-cellular environments; for instance, cardiac 3D-spheroids used to study diseased human heart cells. Importantly, these spheroids further allows the assessment of redox-activity differences between human healthy and dilated myocardium-derived primary mesenchymal cells by scanning electrochemical microscopy, which undoubtedly makes it more convenient to use
[41]. Several approaches have been employed to generate the spheroids.
2.3. Microfluidic Technology
The recent developments in microfluidic technology have made it a tool of great significance in cell culture and assays, with its main application in in vitro 3D-cell culture mimicking the in vivo tissue microenvironment
[42]. It is based on the fabrication of small devices (microsized) having microchannels and chambers which control the behaviour of fluids in them. Microfluidic fabrication in 3D-cell culture, which was first developed in the 1980s, offers a number of advantages, including a controlled cellular microenvironment without intrusions from the outside environment, less reagent ingesting, corresponding processing and analysis, and so on
[43][44]. The small amount of microfluidics for the fabrication of a small number of cells can result in the production of biomimetic models with in vivo tumor microenvironment
[45], different types of cells
[46], and biochemical gradients
[47]. These simple and less costly models provide efficient and high-throughput screening of drugs at the cellular, organ, and whole-body level. The use of microfluidics provides in situ platforms for drug screening with reproducible results
[48]. The use of transparent materials in microfluidic devices can further simplify the direct analysis of various cells based on absorbance or fluorescence
[49] and the direct analysis of tagged proteins in microtissues
[50]. Gelatin
[51] PDMS
[52] are most widely used for the design of new microfluidic devices. For instance, a method of fabricating a 3D cell culture system by stacking multiple layers of PDMS embedded with functionalized hydroxypropyl cellulose methacrylate porous scaffolds was reported, which employed thread as a cost-effective transportation channel to overcome diffusion limitation by continuously supplying nutrients and removing waste
[53].
The analysis of 3D cultures plays a key role in designing and testing types of microfluidic devices. The type of analysis often decides the selection of microfluidic systems, channel volumes, or dilution factors for efficient results
[54]. Bauer et al. reported the use of microfluidic channels with hydrogels for co-culture of breast cancer cells
[55]. In continuation of their work, they successfully analysed the F-actins and tubulin proteins in the 3D-cell culture. The results also demonstrated the biomimetic nature of this 3D culture over 2D culture in mobility edges
[56]. For example, Mosadegh et al. employed the microfluidic device for fluorescent imaging of lung cancer cells for the determination of cell migration under different oxygen gradients. The 3D culture was done on stacked papers and each layer was analysed after 24 h intervals of exposure to gas and media
[57]. Similarly, various drugs or their formulations were also screened for their ADMET properties using microfluidic device-based cell cultures (3D organoids). One such example was the screening of drugs (Cisplatin) on kidney tissue designed using microfluidic devices
[58]. The kidney tissue was created on microfluidic channels and then used to study the filtration, reabsorption, and toxic effects of cisplatin on renal cells and nephrons. Jang et al. designed a multi-layered microfluidic device (PDMS microfluidic channels) for developing biomimicking tubular environments. The rat inner medullary cells were cultured in the system which was observed to transport water-soluble protein within cells like in vivo tissue to control water and ion balance
[59]. The use of microfluidic devices for the development of tissue-on-chip is well explored today for the screening of drug molecules. Lungs-on-chip was developed using microfluidic technology to study the toxicity of drugs, blockage of airways
[60], oxygen transfer efficiency
[61], inflammatory effects on lung cells when exposed to various pathogens or nanoparticles
[62], and other stress factors
[63]. A liver-on-chip model was built to study pharmacokinetic parameters, hepatotoxicity
[64], and phase I/II metabolism of drug molecules
[65]. Deosarkar et al. developed a neonatal blood-brain barrier on-chip model to study biomimetic nature and permeability as well as the human blood-brain barrier
[66]. Jiang et al. created two co-polyester and poly(dimethylsiloxane)-based microfluidic devices for drug molecule screening
[67]. Dhiman et al. reviewed recent developments in on-chip tumor models for combinatorial screening of drug molecules
[68]. Muscle-on-chip is another application of microfluidic technology to provide a small molecule screening platform against a number of muscular dysfunctions such as myasthenia gravis, muscular dystrophy, mitochondrial myopathy, etc.
[69]. All the above-discussed factors describe the key importance of microfluidic technology in 3D culture along with the drug development phase
[70].
3. Role of 3D Cell Culture Models in Drug Repositioning
3D cell cultures have been well established models for drug discovery, disease modelling, drug testing, and toxicity analysis in comparison to conventional 2D models
[71]. Beyond this, the pattern studies of transcriptional factors’ expression and receptor behaviour are implemented by 3D-culture models, which have a great application in drug repositioning and repurposing
[72]. Drug repositioning is a technique that uses the therapeutic value of an existing drug by targeting ailments other than the one for which it was originally approved
[73][74]. A combination of microarrays, bioinformatics, and 3D-culturing models is an excellent approach for drug repositioning
[75]. 3D-cell culture induced gene expression has paved a new path for drug discovery and drug repositioning. A prominent study reported 3D-cell-culture-induced gene expression changes in human neuroblastoma cells by analyzing 1766 genes using microarray analysis performed through RT-PCR reaction
[76]. Some gene expression changes were noted, and it was concluded that several changes in features of cultured cells resulted from varied gene expression. In another study, microarray analysis performed over 9600 genes by smooth muscle cells. It is worth noting that 77 genes were expressed more than twice in 3D-cultured cells. The enhanced cyclin-dependent kinase inhibitor 1 and decreased tyrosine phosphorylation of adhesion kinase observed in 3D-cultured cells were significant indicators of drug repositioning
[77]. Tsunoda et al. reported a potential application of non-malignant prostatic cells in the study of prostate cancer biomarkers beyond the study of genes associated with prostate cancer
[78]. Yin et al. reported the study of multidrug-resistance-hepatotoxic effects of methotrexate in rats along with the multidrug resistance caused by the MRP2 gene
[79]. Pruksakron et al. have studied targets associated with nucleotide metabolism along with mitochondrial and proteins associated with aerobic glycolysis
[80]. Breslin et al. studied differential responses of targets to drugs, modified expression of targets associated with drug resistance in human breast cancer-cell lines along with the study of proteins and enzymes associated with anticancer drugs
[81]. Horning et al. have studied the surface-engineered 3D cultures of breast cancer-cell lines and studied the effects of anticancer drugs on them. Along with this, they studied the significant discrepancy in the action of drugs and the associated factors with it
[82]. In another study, Loessner et al. studied the effects of paclitaxel on bioengineered hydrogel cultures of ovarian cancer epithelial-cell lines along with the study of drug-resistance patterns in the cells
[83]. Nirmalandhan et al. have prepared collagen gel-cell cultures of human lung cancer-cell lines to study the activity of anticancer drugs. They also studied various alterations in the action induced by the drugs
[84].
Recently, a group of researchers fabricated 3D-lung-cancer organoids by using a pleural effusion aspirate and most importantly by incorporating cells obtained from the patients directly to enable personalized disease modelling and tumor characterization
[85]. Interestingly, the isolated patient cells-derived organoids demonstrated anatomically relevant structures and exhibited cancer-specific characteristics that enabled comparative assessment of chemotherapy responses. Another group has reported a 3D cell-based phenotypic assay that determined the effects of radiation and ten established chemotherapeutics in radiation-resistant breast cancer cells grown in 3D-microtissue spheroids
[86]. In the study, heterotypic cultures of normal human dermal fibroblasts and three mammary cancer-cell lines (T47D, MDA-MB-231, and MDA-MB-361) were used to recapitulate the complexity of mammary cancer. Of the ten drugs analysed, vinblastine was found to be more effective, when concurrently given with radiation therapy. A novel in vitro 3D-printed fluidic device that allows nutrient exchange and the diffusion of toxic metabolites from the spheroids to outside was also reported
[87]. Notably, MALDI imaging MS revealed that prodrug irinotecan (a chemotherapeutic agent) penetrated into the tumor spheroids and localized into the center of the spheroid (in the necrotic core), and its metabolite SN-38 was concentrated on the outside region (representing the capability of the cells to metabolize the prodrug). Undoubtedly, this finding supports that the model is efficient enough to mimic the in vivo conditions and can be used to assess drug penetration and metabolism in cancerous cells. A similar finding was reported in 3D-multicellular tumor spheroids used to analyse the distribution of irinotecan as measured by serial trypsinization and nanoflow liquid chromatography-tandem mass spectrometry
[88].
The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), recently wreaked havoc around the world, prompting a push for faster drug discovery and development
[89][90]. Among the different 3D-cell-culture techniques, organoid models (followed by microfluidics-based platform) have been widely considered for COVID-19 research, including alveolar lung organoids, hPSC-derived airway organoids, adult bronchial organoids, hPSC-derived kidney organoids, hPSC-derived liver organoids, brain organoids, etc.. A detailed explanation of these models can be found at
[91][92]. Nevertheless, as compared to the animal models, organoids still exhibit certain limitations due to the lack of blood vessels or vasculature, immune cells, and interorgan communication.
Table 1 represents certain 3D-culture model types and their potential role in drug repositioning.
Table 1. Potential role of 3D cell culture models in drug repositioning.
| Sl. No. |
3D Cell Culture System |
Primary Application |
Application for Drug Repositioning |
Reference |
| 1. |
Micro-dissected tissues of non-malignant prostatic cells |
Prostate cancer associated with RWPE-1 and TA1 genes |
Study of prostate cancer biomarkers |
[78] |
| 2. |
Gel entrapped culture of hepatocytes |
Study of MRP2 gene expression |
Study of multidrug resistance and evaluation of new drug combinations |
[79] |
| 3. |
Collagen-based scaffold culture of HepG2 cell lines |
Proteins of mitochondria and aerobic glycolysis |
Targets in nucleotide metabolism |
[80] |
| 4. |
PolyHEMA scaffold culture of HER2-positive breast cancer cell lines |
Study of anti-cancer drugs, associated proteins and enzymes |
Study of differential responses to drugs, increased expression of targets involved in drug resistance, metabolism |
[81] |
| 5. |
Surface-engineered breast cancer cell lines MCF7 |
Study of action of tamoxifen, doxorubicin, paclitaxel etc. |
Decreased anti-proliferative activity of the drugs |
[82] |
| 6. |
Gel-entrapped culture of human hepatoma cells |
Study of methotrexate |
Study of increased drug resistance and modulation through hormones |
[79] |
| 7. |
Hydrogel matrix of human ovarian cancer cell lines |
Paclitaxel |
Resistance for anticancer action |
[83] |
| 8. |
Collagen gel-based cultures of lung cancer cell lines |
Paclitaxel, doxorubicin, cisplatin, gemcitabine |
Alterations in drug-induced activity |
[84] |