Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carmen Lopez-Sanchez | -- | 2012 | 2022-08-08 11:04:54 | | | |
| 2 | Conner Chen | Meta information modification | 2012 | 2022-08-09 08:06:54 | | | | |
| 3 | Conner Chen | Meta information modification | 2012 | 2022-08-10 08:29:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Garcia-Padilla, C.; Lozano-Velasco, E.; Muñoz-Gallardo, M.D.M.; Castillo-Casas, J.M.; Caño-Carrillo, S.; Martínez-Amaro, F.J.; García-López, V.; Aránega, A.; Franco, D.; García-Martínez, V.; et al. LncRNA H19 Impairs Chemo and Radiotherapy in BC. Encyclopedia. Available online: https://encyclopedia.pub/entry/25947 (accessed on 07 February 2026).
Garcia-Padilla C, Lozano-Velasco E, Muñoz-Gallardo MDM, Castillo-Casas JM, Caño-Carrillo S, Martínez-Amaro FJ, et al. LncRNA H19 Impairs Chemo and Radiotherapy in BC. Encyclopedia. Available at: https://encyclopedia.pub/entry/25947. Accessed February 07, 2026.
Garcia-Padilla, Carlos, Estefanía Lozano-Velasco, María Del Mar Muñoz-Gallardo, Juan Manuel Castillo-Casas, Sheila Caño-Carrillo, Francisco José Martínez-Amaro, Virginio García-López, Amelia Aránega, Diego Franco, Virginio García-Martínez, et al. "LncRNA H19 Impairs Chemo and Radiotherapy in BC" Encyclopedia, https://encyclopedia.pub/entry/25947 (accessed February 07, 2026).
Garcia-Padilla, C., Lozano-Velasco, E., Muñoz-Gallardo, M.D.M., Castillo-Casas, J.M., Caño-Carrillo, S., Martínez-Amaro, F.J., García-López, V., Aránega, A., Franco, D., García-Martínez, V., & López-Sánchez, C. (2022, August 08). LncRNA H19 Impairs Chemo and Radiotherapy in BC. In Encyclopedia. https://encyclopedia.pub/entry/25947
Garcia-Padilla, Carlos, et al. "LncRNA H19 Impairs Chemo and Radiotherapy in BC." Encyclopedia. Web. 08 August, 2022.
Copy Citation
Long non-coding RNAs, H19 in particular, have been revealed as powerful protective factors in various types of cancer, including breast cancer (BC).
lncRNA H19
chemo-resistance
radio-resistance
1. Introduction
For decades, scientists have considered non-coding RNAs (ncRNAs) as a non-functional part of the genome, focusing their attention primarily on coding RNA biology. The sequencing of the human genome and later the ENCODE project have shown that more than 80% of the genome is transcribed in some type of RNA. Interestingly, only 3% of this transcribed genome corresponds to coding RNAs, suggesting that ncRNAs are as significant or more significant than coding RNAs [1][2]. It has been demonstrated that non-coding RNAs are essential for the regulation of cellular pathways and biological processes such as cell development, differentiation, growth, and homeostasis, as well as diseases [3][4][5][6].
According to their length, ncRNAs can be classified into: (i) small non-coding RNAs, with less than 200 nucleotides, including microRNAs, snoRNAs, piRNAs, and tRNAs [7]; and (ii) long non-coding RNAs (lncRNAs) with more than 200 nucleotides, including intronic lncRNAs, enhancer lncRNAs, circular lncRNAs, and intergenic lncRNAs [8].
With respect to microRNAs, these present 20–22 ribonucleotides in length on average and display the capacity to bind to the 3′ untranslated region (3′UTR) of coding RNAs by complementary base pairing, promoting their degradation and/or translational blockage. The role of microRNAs as post-transcriptional modulators has been widely described in multiple biological and cellular processes, including cell development, differentiation, growth, and homeostasis, as well as diseases [9][10][11][12].
In the case of lncRNAs, these are structurally similar to mRNAs since they are transcribed by RNA polymerase II, and have the same typical post-transcriptional modifications, in 5′ terminal cap and 3′ terminal poly (A) in particular. Notably, they lack the capacity to code proteins. Mechanistically, lncRNAs can act both as transcriptional regulators (by modulation of nuclear gene expression in different ways, including epigenetic landscape control, transcriptional complex scaffolding, and decoy molecules) and as post-transcriptional regulators (modulating microRNA degradation, mRNA stability, and/or protein translation). A deeper study of these lncRNAs roles will help people to better understand the regulation of multiple biological processes [13][14][15][16].
A particular lncRNA, H19 (produced by H19 gene), is abundantly expressed during embryonic development, mainly in derived tissues from the endoderm and mesoderm, and downregulated after birth, except for muscle tissues, such as skeletal and cardiac muscles [17][18][19]. H19 gene is highly conserved between primates and rodents, maintaining both structure and expression pattern in different species. However, it shows substantial differences in terms of the putative ORFs (open reading frames) arrangement present in this gene, which exhibits 5 exons separated by 4 introns, structurally [20]. While in humans a maximum of 13 different isoforms are observed, only 7 are identified in mice. H19 gene generates a 2.3 kb transcript with a cap in 5′ end and a polyA tail in 3′ end [21]. Subcellular expression of H19 depends on cellular type and biological context, showing its expression both in the cytoplasm and nucleus [22][23]. In the genome, H19 gene is located within a locus highly regulated by epigenomic machinery. This locus, named H19-Igf2, showing several differentially methylated regions (DMRs) and imprinting control regions (ICRs), leads to intensive modulation of genes contained in it, depending on the process and/or biological context [24][25]. Interestingly, H19 encodes the primary microRNA precursor for miR-675. The expression of this microRNA is not dependent on RNA H19 transcription, although both expressions are correlated in several biological processes [26][27]. The H19 gene has been studied for some time now, although its function is still not well defined. In particular, it acts as a trans-regulator of a group of co-expressed genes as part of an imprinted network, which is likely to control cellular homeostasis [24][25][26][27]
Taking into account that non-coding RNA genome has proved to play a pivotal role, both as tumor suppressor and oncogenic molecules in cancer therapy [28][29][30][31][32], it is critical to understand the underlying molecular mechanisms in favor of new therapeutic approaches for cancer treatment efficiency. Since the resistance to chemo and radiotherapy constitutes a crucial factor involved in disease relapse and metastasis [33][34], it is of great importance to establish the intrinsic factors involved in this process. Known molecular mechanisms of chemo and radioresistance include transporter pumps, oncogenes, tumor suppressor gene, mitochondrial alteration, DNA repair, autophagy, epithelial–mesenchymal transition (EMT), cancer stemness, and exosome and extensive epigenetic regulation, among others [35][36][37][38]. Noticeably, abnormal expression of lncRNA H19 has been found in different types of tumor cells, affecting cancer progression through different mechanisms, either as a suppressor or oncogenic gene, depending on cellular context [39][40][41][42][43][44][45]. For this reason, clinically, lncRNA H19 could be useful as a biomarker of diagnosis, therapy, and prognosis in cancer [46][47]. In this sense, those clear-cut mechanisms underlying the H19 regulatory roles in the biological progression of cancer requires further investigation.
2. LncRNA H19 Impairs Chemo and Radiotherapy in Breast Cancer
Breast cancer is the most frequently diagnosed malignancy and the second leading cause of cancer mortality in females worldwide [48]. Broadly, breast cancer patients can be classified into estrogen receptor positive (ER+)—corresponding to 70% of total cases—or estrogen receptor negative (ER−) [49][50]. Drug treatment against both types of breast cancer differs from each other, since several drugs are antagonists of estrogen receptors and exerting their functions blocking binding between ER and estrogens. This binding leads to disrupting the activation of downstream signaling pathway dependent of RE-estrogens interaction [51]. Currently, administration of three drugs, tamoxifen (TMX), doxorubicin (DOX), and paclitaxel (PTX), are considered as first-line of chemotherapy [52]. The application of any of these drugs separately or in combination with different types of drugs that enhance their cytotoxicity against malignant cells, considerably improves the clinical outcome of breast cancer, even though their continuous use results in acquired resistance of breast cancer cells against drug treatment [53][54][55]. To reverse such resistance and re-sensitize cells to different drugs, it is vitally important to understand the underlying molecular mechanisms that modulate their resistance acquisition. Several studies have pointed out the role of H19 as a powerful oncogenic agent in this process, pinpointing it as a critical modulator of chemoresistance (Figure 1).
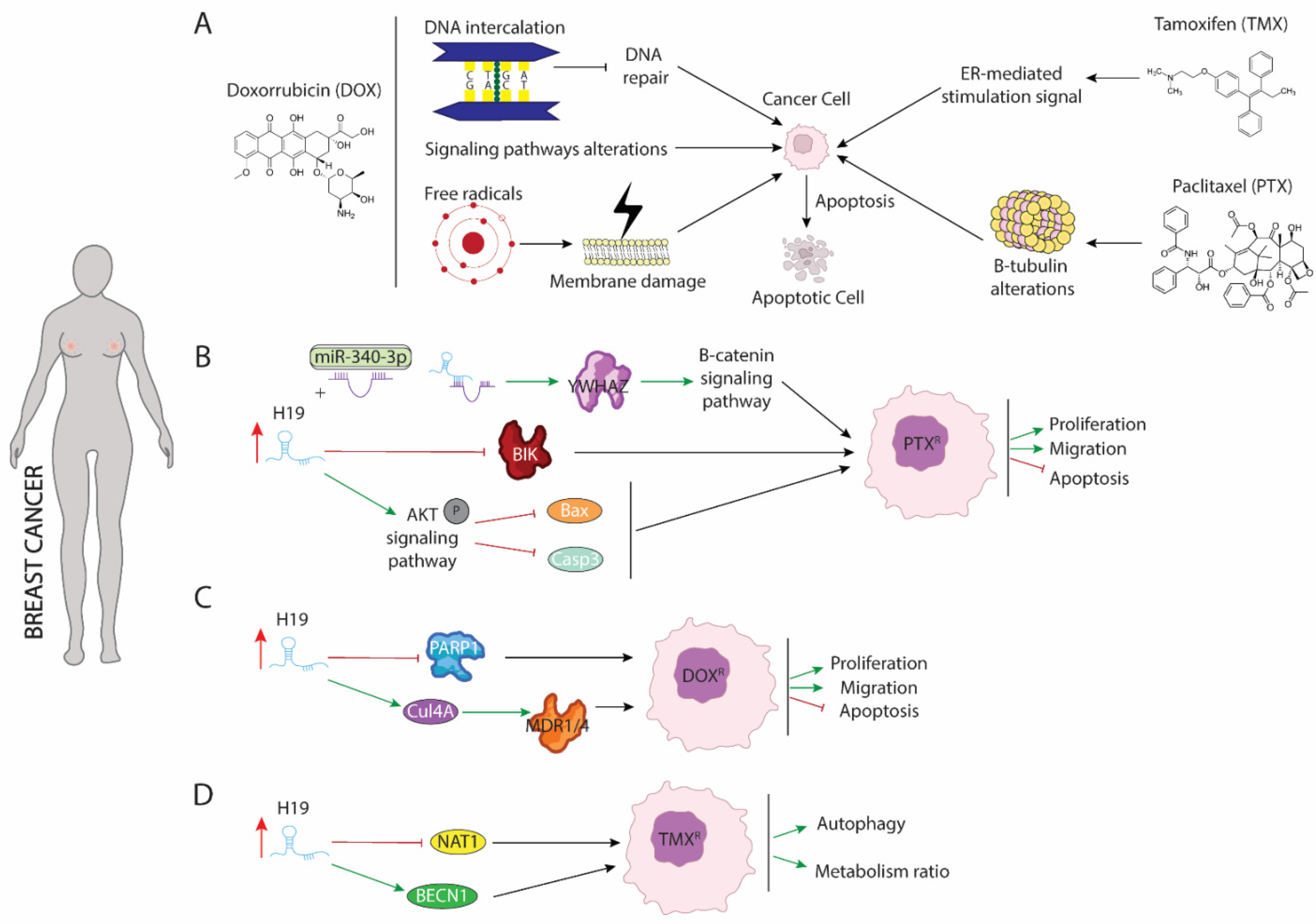
Figure 1. Schematic representation of molecular mechanism dependent of H19 function in breast cancer. (A) Modes of action of paclitaxel (PTX), doxorubicin (DOX), and tamoxifen (TMX) drugs involved in breast cancer treatment. (B) Role of H19 as a sponge to binding miR-340 enhancing expression of YWHAZ protein and increasing proliferation and migration while blocking apoptosis. (C) H19 increase expression of CUL4A and repress expression of PARP1 lead to DOX resistance in breast cancer. (D) H19 modulates methylation of BECN1 promotor and blocking NAT1 expression enhancing autophagy and metabolism procedures.
Paclitaxel (PTX) is the most commonly used antitumor treatment drug on several types of carcinomas [55]. Mechanistically, PTX action includes several signaling pathways in which PTX modulates cellular processes that results in programmed cell death triggering. PTX is considered as the first-line treatment drug in breast cancer (BC), especially in triple negative breast cancer (TNBC) [56], since this subtype of carcinoma is not sensitive to estrogen receptor positive breast cancer cell. Unfortunately, the resistance of BC to PTX treatment is a great obstacle in clinical applications and one of the major causes of death associated with treatment failure. In breast cancer, PTX (Figure 1A) induces cellular apoptosis by interacting with β-tubulin and altering microtubules stability. PTX-resistance is acquired by mutations in α or β-tubulin, enhancing p53 and AKT activation signaling pathways or by dysregulation of apoptotic proteins [57][58]. Functional assays in both ER(+)-MCF-7 and ER(-)-Triple negative breast cancer (TNBC) carcinomas have shown that acquisition of resistance to PTX requires the upregulation of H19 (Figure 1B), which in turn blocks activation of several apoptotic pathways. Curiously, in ERα + breast cancer, H19 modulates resistance of PTX at both transcriptional and post-transcriptional levels. Si et al. (2016) [59] demonstrated that upregulation of H19 in MCF-7 cell line inhibited transcription of BCL-2 interacting killer protein (BIK)—a proapoptotic BH3-only member of the BCL-2 family which is prognostic for relapse and decreased overall survival of breast cancer [60]—by recruiting EZH2 subunit to the promoter of this gene, modulating H3 methylation at lysine 27. As result of BIK promoter methylation, PTX-resistance MCF-7 cells display increased proliferation rate and decreased cellular apoptosis [59]. Furthermore, H19 blocks miR-340-3p function—a known tumor suppressor miRNA involved in repression of EMT—acting as competitive sponge and avoiding degradation of tyrosine 3-monooxygenase/tryptophan 5-monoixygenase activation protein (YWHAZ), which in turn enhances activation of Wnt/β-catenin signaling pathway. As results of increased activity of YWHAZ-Wnt/β-catenin axis, PTX-resistance cells exhibit increased metastasis, invasion, and EMT. Additionally, the phenotype associated with H19 upregulation is recapitulated in both xenograft models and biopsies from patients with PTX resistance [61]. Unlike ER(+), increased H19 levels in TNBC promote activation of AKT by phosphorylation. As a consequence of enhancing AKT activity, BAX and cleaved caspase 3 are repressed, and cellular apoptosis is inhibited. Furthermore, downregulation of H19 in TNBC is translated into lower tumor growth rate in vivo and reduced cell proliferation accompanied by higher rates of apoptosis [62].
Doxorubicin (DOX) is a member of the anthracycline family and currently is one of the main treatments of choice for the treatment of both ER+ and ER- breast cancer [54]. DOX negatively affects the survival of malignant cells through different mechanisms of action (Figure 1A): (1) DOX is capable of mediating intercalation into DNA and disruption of topoisomesare-II function, avoiding DNA repair, and increasing cellular apoptosis; (2) DOX promotes generation of free radicals and their damage to cellular membranes, proteins, and DNA; (3) DOX deregulates several pivotal pathways involved in tumorogenesis such as PI3K/mTOR/AKT or ERK signaling [63]. Several studies have pointed out H19 as a major mediator of DOX chemoresistance (Figure 1C) by modulating MDR1/MDR4 and PARP1 expression [64][65]. Increased expression of H19 is required for the acquisition of DOX-resistance in several breast cancer cell lines. Curiously, downregulation of H19 reduces cell viability, lowers colony forming, and increases apoptosis under DOX treatment. Mechanistically, H19 promotes expression of Cullin 4A (CUL4A), a ubiquitin ligase component [66], which in turn enhances expression of ABCB1/4 genes that encoded MDR1/4 proteins, two members of the ATP-binding cassette family highly upregulated in several carcinomas, including breast cancer [67][68]. Both proteins exert pivotal functions in oncogenesis acting as inductors of multidrug resistance [69]. However, the molecular mechanisms dependent on MRD1/MDR4 are still unclear and require additional and intensive investigations. Wang et al. (2020) [65] showed that H19-induced DOX-chemoresistance is mediated by repression of PARP1. In MCF-7, high levels of H19 were correlated with downregulated PARP1 expression. Even though PARP1 is upregulated in several types of carcinomas and PARP1 inhibitors have been described as pivotal drugs against tumorigenesis, PARP1 has been shown to increase the antitumor activity of other drugs such as temozolomide and topotecan in preclinical studies, including models of pediatric cancers [70][71][72]. Curiously, it is capable of being packaged into exosomes from resistance cells and this diffusion towards sensitive cells leads to the acquisition of resistance against DOX treatment [73].
Tamoxifen (TMX), an anti-estrogen (Figure 1A), competitively inhibits estrogen binding to the ER and blocks the ER-mediated stimulation signal [53]. Five years of tamoxifen adjuvant therapy has been shown to safely reduce 15-year risk of breast cancer recurrence and death; however, a substantial group of patients was shown to eventually develop resistance (de novo or acquired) to tamoxifen. Although many molecular mechanisms of tamoxifen resistance have been described, including mutations in the ESR1 gene and the activation of alternative growth pathways, such as ERBB2/HER2, EGFR, IGF1R, and cyclin D1/CDK4/6 pathways, it remains necessary to gain an improved understanding of the potential mechanisms of tamoxifen resistance [74][75][76][77]. Several studies have pointed out the importance of upregulation of H19 in TMX resistance acquisition. Increased level of H19 in TMX-resistance cells (Figure 1D) is translated into enhanced autophagy activity and higher metabolism ratio by upregulation of Beclin1 (BECN1) and downregulation of N-acetyltransferase-1 (NAT1) transcription, respectively [78][79]. H19 downregulates the methylation state of Beclin1 promoter by binding and inhibiting S-adenosyl homocysteine hydrolase (SAHH). As consequence, DNMT3B function is reduced, resulting in lower ratio of methylation of Beclin1 promoter and thus higher Beclin1 expression [78]. Additionally, H19 modulate methylation of NAT1 promoter reducing transcription of it. NAT1 exert a pivotal role in metabolism of carcinogens and it is negatively correlated with a poor prognosis and aggressiveness of ER(+) breast cancer [79].
References
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563.
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774.
- García-Padilla, C.; Aránega, A.; Franco, D. The role of long non-coding RNAs in cardiac development and disease. AIMS Genet. 2018, 5, 124–140.
- Expósito-Villén, A.; Aránega, A.E.; Franco, D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition. Non-Coding RNA 2018, 4, 14.
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166.
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992.
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933.
- Singh, S.R.; Rameshwar, P. MicroRNA in Development and in the Progression of Cancer; Springer: New York, NY, USA, 2014.
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An Analysis of Human MicroRNA and Disease Associations. PLoS ONE 2008, 3, e3420.
- Garcia-Padilla, C.; Garcia-Lopez, V.; Aranega, A.; Franco, D.; Garcia-Martinez, V.; Lopez-Sanchez, C. Inhibition of RhoA and Cdc42 by miR-133a Modulates Retinoic Acid Signalling during Early Development of Posterior Cardiac Tube Segment. Int. J. Mol. Sci. 2022, 23, 4179.
- Garcia-Padilla, C.; Dueñas, A.; Franco, D.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V.; Lopez-Sanchez, C. Dynamic MicroRNA Expression Profiles During Embryonic Development Provide Novel Insights into Cardiac Sinus Venosus/Inflow Tract Differentiation. Front. Cell Dev. Biol. 2022, 9, 767954.
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nat. Cell Biol. 2016, 539, 452–455.
- Mathieu, E.L.; Belhocine, M.; Dao, L.T.; Puthier, D.; Spicuglia, S. Functions of lncRNA in development and diseases. Med. Sci. 2014, 30, 790–796. (In French)
- García-Padilla, C.; Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Jiménez-Sábado, V.; Ginel, A.; Hove-Madsen, L.; Aránega, A.E.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2022, 36, e22051.
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yuyang, L.J.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652.
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36.
- Milligan, L.; Forné, T.; Antoine, E.; Weber, M.; Hémonnot, B.; Dandolo, L.; Brunel, C.; Cathala, G. Turnover of primary transcripts is a major step in the regulation of mouse H19 gene expression. EMBO Rep. 2002, 3, 774–779.
- Schoenfelder, S.; Smits, G.; Fraser, P.; Reik, W.; Paro, R. Non-coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep. 2007, 8, 1068–1073.
- Zemel, S.; Bartolomei, M.S.; Tilghman, S.M. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 1992, 2, 61–65.
- García-Padilla, C.; Domínguez, J.N.; Aránega, A.E.; Franco, D. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194435.
- Wei, Y.; Liu, Z.; Fang, J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let-7c in gastric cancer. Mol. Med. Rep. 2018, 17, 2600–2606.
- Viereck, J.; Bührke, A.; Foinquinos, A.; Chatterjee, S.; Kleeberger, J.A.; Xiao, K.; Janssen-Peters, H.; Batkai, S.; Ramanujam, D.; Kraft, T.; et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur. Heart J. 2020, 41, 3462–3474.
- Charalambous, M.; Menheniott, T.R.; Bennett, W.R.; Kelly, S.M.; Dell, G.; Dandolo, L.; Ward, A. An enhancer element at the Igf2/H19 locus drives gene expression in both imprinted and non-imprinted tissues. Dev. Biol. 2004, 271, 488–497.
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480.
- Lv, J.; Wang, L.; Zhang, J.; Lin, R.; Wang, L.; Sun, W.; Wu, H.; Xin, S. Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem. Biophys. Res. Commun. 2018, 497, 1154–1161.
- Luo, H.; Wang, J.; Liu, D.; Zang, S.; Ma, N.; Zhao, L.; Zhang, L.; Zhang, X.; Qiao, C. The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARα. Mol. Immunol. 2019, 105, 46–54.
- Zhu, L.; Zhu, Y.; Han, S.; Chen, M.; Song, P.; Dai, D.; Xu, W.; Jiang, T.; Feng, L.; Shin, V.Y.; et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019, 10, 383.
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026.
- Luo, Y.; Zheng, S.; Wu, Q.; Wu, J.; Zhou, R.; Wang, C.; Wu, Z.; Rong, X.; Huang, N.; Sun, L.; et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 2021, 17, 4083–4101.
- Han, P.; Li, J.W.; Zhang, B.M.; Lv, J.C.; Li, Y.M.; Gu, X.Y.; Yu, Z.W.; Jia, Y.H.; Bai, X.F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9.
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019, 38, 4637–4654.
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348.
- Wu, P.; Gao, W.; Su, M.; Nice, E.C.; Zhang, W.; Lin, J.; Xie, N. Adaptive Mechanisms of Tumor Therapy Resistance Driven by Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 641469.
- Zheng, H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017, 8, 59950–59964.
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The Role of Notch3 Signaling in Cancer Stemness and Chemoresistance: Molecular Mechanisms and Targeting Strategies. Front. Mol. Biosci. 2021, 8, 694141.
- Zhang, F.; Wang, H.; Yu, J.; Yao, X.; Yang, S.; Li, W.; Xu, L.; Zhao, L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer 2021, 20, 6.
- Käsmann, L.; Dietrich, A.; Staab-Weijnitz, C.A.; Manapov, F.; Behr, J.; Rimner, A.; Jeremic, B.; Senan, S.; De Ruysscher, D.; Lauber, K.; et al. Radiation-induced lung toxicity—Cellular and molecular mechanisms of pathogenesis, management, and literature review. Radiat. Oncol. 2020, 15, 214.
- Gan, L.; Lv, L.; Liao, S. Long non-coding RNA H19 regulates cell growth and metastasis via the miR-22-3p/Snail1 axis in gastric cancer. Int. J. Oncol. 2019, 54, 2157–2168.
- Wang, J.; Sun, J.; Yang, F. The role of long non-coding RNA H19 in breast cancer. Oncol. Lett. 2020, 19, 7–16.
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824.
- Huang, Z.; Chu, L.; Liang, J.; Tan, X.; Wang, Y.; Wen, J.; Chen, J.; Wu, Y.; Liu, S.; Liao, J.; et al. H19 Promotes HCC Bone Metastasis Through Reducing Osteoprotegerin Expression in a Protein Phosphatase 1 Catalytic Subunit Alpha/p38 Mitogen-Activated Protein Kinase-Dependent Manner and Sponging microRNA 200b-3p. Hepatology 2021, 74, 214–232.
- Xu, H.; Ding, Y.; Yang, X. Overexpression of Long Noncoding RNA H19 Downregulates miR-140-5p and Activates PI3K/AKT Signaling Pathway to Promote Invasion, Migration and Epithelial-Mesenchymal Transition of Ovarian Cancer Cells. Biomed. Res. Int. 2021, 2021, 6619730.
- Liu, Z.Z.; Tian, Y.F.; Wu, H.; Ouyang, S.Y.; Kuang, W.L. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma 2020, 67, 111–118.
- Liu, C.; Chen, Z.; Fang, J.; Xu, A.; Zhang, W.; Wang, Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016, 37, 263–270.
- Shermane Lim, Y.W.; Xiang, X.; Garg, M.; Le, M.T.; Li-Ann Wong, A.; Wang, L.; Goh, B.C. The double-edged sword of H19 lncRNA: Insights into cancer therapy. Cancer Lett. 2021, 500, 253–262.
- Alipoor, B.; Parvar, S.N.; Sabati, Z.; Ghaedi, H.; Ghasemi, H. An updated review of the H19 lncRNA in human cancer: Molecular mechanism and diagnostic and therapeutic importance. Mol. Biol. Rep. 2020, 47, 6357–6374.
- Katsura, C.; Ogunmwonyi, I.; Kankam, H.K.; Saha, S. Breast cancer: Presentation, investigation and management. Br. J. Hosp. Med. 2022, 83, 1–7.
- Arciero, C.A.; Guo, Y.; Jiang, R.; Behera, M.; O’Regan, R.; Peng, L.; Li, X. ER+/HER2+ Breast Cancer Has Different Metastatic Patterns and Better Survival Than ER-/HER2+ Breast Cancer. Clin. Breast Cancer 2019, 19, 236–245.
- Keenan, T.E.; Tolaney, S.M. Role of Immunotherapy in Triple-Negative Breast Cancer. J. Natl. Compr. Canc. Netw. 2020, 18, 479–489.
- Belachew, E.B.; Sewasew, D.T. Molecular Mechanisms of Endocrine Resistance in Estrogen-Positive Breast Cancer. Front. Endocrinol. 2021, 12, 599586.
- Fisusi, F.A.; Akala, E.O. Drug Combinations in Breast Cancer Therapy. Pharm. Nanotechnol. 2019, 7, 3–23.
- Mills, J.N.; Rutkovsky, A.C.; Giordano, A. Mechanisms of resistance in estrogen receptor positive breast cancer: Overcoming resistance to tamoxifen/aromatase inhibitors. Curr. Opin. Pharmacol. 2018, 41, 59–65.
- Shafei, A.; El-Bakly, W.; Sobhy, A.; Wagdy, O.; Reda, A.; Aboelenin, O.; Marzouk, A.; El Habak, K.; Mostafa, R.; Ali, M.A.; et al. A review on the efficacy and toxicity of different doxorubicin nanoparticles for targeted therapy in metastatic breast cancer. Biomed. Pharmacother. 2017, 95, 1209–1218.
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789.
- Hsu, M.Y.; Hsieh, C.H.; Huang, Y.T.; Chu, S.Y.; Chen, C.M.; Lee, W.J.; Liu, S.J. Enhanced Paclitaxel Efficacy to Suppress Triple-Negative Breast Cancer Progression Using Metronomic Chemotherapy with a Controlled Release System of Electrospun Poly-d-l-Lactide-Co-Glycolide (PLGA) Nanofibers. Cancers 2021, 13, 3350.
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer. 2010, 10, 194–204, Erratum in Nat. Rev. Cancer 2010, 10, 309.
- Jabbarzadeh Kaboli, P.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806.
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462.
- Pandya, V.; Glubrecht, D.; Vos, L.; Hanson, J.; Damaraju, S.; Mackey, J.; Hugh, J.; Goping, I.S. The pro-apoptotic paradox: The BH3-only protein Bcl-2 interacting killer (Bik) is prognostic for unfavorable outcomes in breast cancer. Oncotarget 2016, 7, 33272–33285.
- Yan, L.; Yang, S.; Yue, C.X.; Wei, X.Y.; Peng, W.; Dong, Z.Y.; Xu, H.N.; Chen, S.L.; Wang, W.R.; Chen, C.J.; et al. Long noncoding RNA H19 acts as a miR-340-3p sponge to promote epithelial-mesenchymal transition by regulating YWHAZ expression in paclitaxel-resistant breast cancer cells. Environ. Toxicol. 2020, 35, 1015–1028.
- Han, J.; Han, B.; Wu, X.; Hao, J.; Dong, X.; Shen, Q.; Pang, H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol. Appl. Pharmacol. 2018, 359, 55–61.
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharm. Genom. 2011, 21, 440–446.
- Zhu, Q.N.; Wang, G.; Guo, Y.; Peng, Y.; Zhang, R.; Deng, J.L.; Li, Z.X.; Zhu, Y.S. LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget 2017, 8, 91990–92003.
- Wang, Y.; Zhou, P.; Li, P.; Yang, F.; Gao, X.Q. Long non-coding RNA H19 regulates proliferation and doxorubicin resistance in MCF-7 cells by targeting PARP1. Bioengineered 2020, 11, 536–546.
- Saucedo-Cuevas, L.P.; Ruppen, I.; Ximénez-Embún, P.; Domingo, S.; Gayarre, J.; Muñoz, J.; Silva, J.M.; García, M.J.; Benítez, J. CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response. Oncotarget 2014, 5, 2330–2343.
- Haque, A.; Sait, K.H.W.; Alam, Q.; Alam, M.Z.; Anfinan, N.; Wali, A.W.N.; Rasool, M. MDR1 Gene Polymorphisms and Its Association with Expression as a Clinical Relevance in Terms of Response to Chemotherapy and Prognosis in Ovarian Cancer. Front. Genet. 2020, 11, 516.
- Yang, M.; Li, H.; Li, Y.; Ruan, Y.; Quan, C. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018, 17, 6211–6226.
- Martin, H.L.; Smith, L.; Tomlinson, D.C. Multidrug-resistant breast cancer: Current perspectives. Breast Cancer 2014, 6, 1–13.
- Pignochino, Y.; Capozzi, F.; D’Ambrosio, L.; Dell’Aglio, C.; Basiricò, M.; Canta, M.; Lorenzato, A.; Vignolo Lutati, F.; Aliberti, S.; Palesandro, E.; et al. PARP1 expression drives the synergistic antitumor activity of trabectedin and PARP1 inhibitors in sarcoma preclinical models. Mol. Cancer 2017, 16, 86.
- Weaver, A.N.; Yang, E.S. Beyond DNA Repair: Additional Functions of PARP-1 in Cancer. Front. Oncol. 2013, 3, 290.
- Takagi, M.; Ogawa, C.; Aoki-Nogami, Y.; Iehara, T.; Ishibashi, E.; Imai, M.; Kihara, T.; Nobori, K.; Hasebe, K.; Mizutani, S.; et al. Phase I clinical study of oral olaparib in pediatric patients with refractory solid tumors: Study protocol. BMC Pediatrics 2019, 19, 31.
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell Physiol. 2020, 235, 6896–6904.
- Elledge, R.M.; Lock-Lim, S.; Allred, D.C.; Hilsenbeck, S.G.; Cordner, L. p53 mutation and tamoxifen resistance in breast cancer. Clin. Cancer Res. 1995, 1, 1203–1208.
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85.
- Riggins, R.B.; Schrecengost, R.S.; Guerrero, M.S.; Bouton, A.H. Pathways to tamoxifen resistance. Cancer Lett. 2007, 256, 1–24.
- Zhu, Y.; Liu, Y.; Zhang, C.; Chu, J.; Wu, Y.; Li, Y.; Liu, J.; Li, Q.; Li, S.; Shi, Q.; et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018, 9, 1595.
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019, 12, 81.
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Oncol. Targets Ther. 2018, 11, 8001–8012.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
613
Revisions:
3 times
(View History)
Update Date:
10 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




