Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mario García-Domínguez | -- | 2642 | 2022-07-25 09:41:30 | | | |
| 2 | Camila Xu | Meta information modification | 2642 | 2022-07-25 09:49:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lara-Ureña, N.; Jafari, V.; García-Domínguez, M. The Sumoylation Pathway. Encyclopedia. Available online: https://encyclopedia.pub/entry/25481 (accessed on 03 March 2026).
Lara-Ureña N, Jafari V, García-Domínguez M. The Sumoylation Pathway. Encyclopedia. Available at: https://encyclopedia.pub/entry/25481. Accessed March 03, 2026.
Lara-Ureña, Nieves, Vahid Jafari, Mario García-Domínguez. "The Sumoylation Pathway" Encyclopedia, https://encyclopedia.pub/entry/25481 (accessed March 03, 2026).
Lara-Ureña, N., Jafari, V., & García-Domínguez, M. (2022, July 25). The Sumoylation Pathway. In Encyclopedia. https://encyclopedia.pub/entry/25481
Lara-Ureña, Nieves, et al. "The Sumoylation Pathway." Encyclopedia. Web. 25 July, 2022.
Copy Citation
Post-translational modifications (PTMs) are key regulators of most biological processes. Besides phosphorylation, methylation, acetylation and others, covalent modification of proteins by small polypeptides of the ubiquitin-like modifiers (UBLs) family have gained importance. Among UBLs, the small ubiquitin-like modifier (SUMO), of ~90 amino acids and discovered in the nineties, has proven to regulate most cellular processes. The sumoylation pathway is quite similar to the ubiquitination pathway, but there is its own set of enzymes for modification by SUMO.
SUMO
protease
ligase
cancer
regulation
1. Introduction
Post-translational modifications (PTMs) are key regulators of most biological processes. Besides phosphorylation, methylation, acetylation and others, covalent modification of proteins by small polypeptides of the ubiquitin-like modifiers (UBLs) family have gained importance in recent decades. Among UBLs, the small ubiquitin-like modifier (SUMO), of ~90 amino acids and discovered in the nineties, has proven to regulate most cellular processes [1].
Up to five SUMO paralogs have been described in vertebrates. While SUMO4–5 present restricted patterns of expression and it is not clear whether they can be functionally conjugated to proteins [2], SUMO1-3 are widely expressed in all tissues and involved in modification of thousands of proteins [3][4][5]. SUMO2 and SUMO3 are virtually indistinguishable and normally designated as SUMO2/3. They are abundant in the cell in the unconjugated form and rapidly attached to proteins in response to a variety of stress stimuli [6][7][8]. By contrast, most SUMO1 is conjugated to proteins, and mainly to the nuclear pore protein RanGAP1 [9][10]. SUMO1 shares about 17% and 50% identity with ubiquitin and SUMO2/3, respectively. SUMO2 and SUMO3 are 97% identical. Sumoylation occurs at the ε-amino group of a Lys (K) residue, often included in the consensus I/L/VKxE/D. SUMO2/3 displays this consensus sequence, which facilitates the formation of poly-SUMO chains.
Despite being able to covalently modify other proteins, SUMO is also able to non-covalently interact with many proteins through SUMO interacting motifs (SIMs) present in interactors [11][12]. SIMs are of special relevance for sumoylation-dependent ubiquitination, through the action of SUMO-targeted ubiquitin ligases (STUbLs) like RNF4, which present tandem SIMs able to recognize poly-SUMO chains in ubiquitin targets to be degraded [13]. The combination of sumoylation sites with SIMs in the same or different proteins contributes to the formation of protein macrostructures which may enhance sumoylation by recruiting additional SUMO targets to a sumoylation favorable environment. For instance, the promyelocytic leukemia protein (PML) is deeply modified by SUMO, which in turn interacts with SIMs in PML giving rise to big aggregates (PML nuclear bodies (NBs)) recruiting additional sumoylated proteins and serving as a sumoylation platform for many other SUMO targets [14]. In this research, SUMO has been considered a molecular glue, and this is related to the concept of “protein group sumoylation”. This refers to the fact that sumoylation, in contrast to many other PTMs, frequently affects collectively to different proteins in a group of interacting proteins, rather than individually to a specific protein [15].
An enigmatic and premature observation on sumoylation is related to the fact that at any given time, only a very reduced fraction of the pool of a target protein appears modified by SUMO, while evidence suggests that the whole target pool is modified. For instance, the use of sumoylation mutants has frequently dramatic consequences, which is not expected if most of the molecules of a given protein make their function in the unmodified form and only a small percentage of molecules is modified for additional purposes. This paradox has been explained on the basis of sumoylation being, in most of the cases, a quite transient modification, but with permanent effects on protein function or destiny once SUMO has been removed [16].
Sumoylation is essential in vertebrates, and different knock out (KO) animal models support this. The unique conjugating enzyme of the sumoylation pathway (UBC9, see next section) has been demonstrated to be indispensable for survival of the mouse embryo. Ubc9 KO mice embryos dye at the early post-implantation stage due to the inability of the blastocyst inner cell mass to expand, which enters in apoptosis [17]. Regarding SUMO paralogs, it has been described that both SUMO1 and SUMO3 are dispensable, probably due to compensation by the other SUMO molecules (reviewed in [18]). However, loss of SUMO2 cannot be compensated by any paralog, which seems to be related to the high abundance of SUMO2 in comparison with the other SUMO molecules [19].
SUMO attachment to proteins has a great impact on their functions, as it may alter localization, activity, stability, interactions and conformation of target proteins. SUMO is involved in regulation of most relevant cellular processes, and in particular in gene expression [20]. Thus, unbalanced sumoylation may lead to altered protein function or gene expression, resulting in cell transformation and tumorigenesis. Key regulators of the sumoylation process are the SUMO ligases and proteases, which determine the sumoylation status of target proteins.
2. The Sumoylation Pathway
The sumoylation pathway is quite similar to the ubiquitination pathway, but there is its own set of enzymes for modification by SUMO (Figure 1). This involves several steps: (i) initial maturation of the SUMO precursor, by proteolysis of several C-terminal amino acids to expose a Gly-Gly (GG) motif; (ii) activation of mature SUMO by the heterodimeric SAE1/UBA2 E1 enzyme, through ATP hydrolysis; and (iii) transfer to UBC9, the unique conjugating E2 enzyme in the sumoylation system. Both activation and transfer to UBC9 involves the formation of a thioester bond between the C-terminus of mature SUMO and a Cys (C) residue in the catalytic sites of E1 or E2, (iv) transfer from UBC9 to targets, frequently assisted by a SUMO ligase or E3. Besides, specific SUMO proteases are in charge of SUMO maturation and scission from targets.
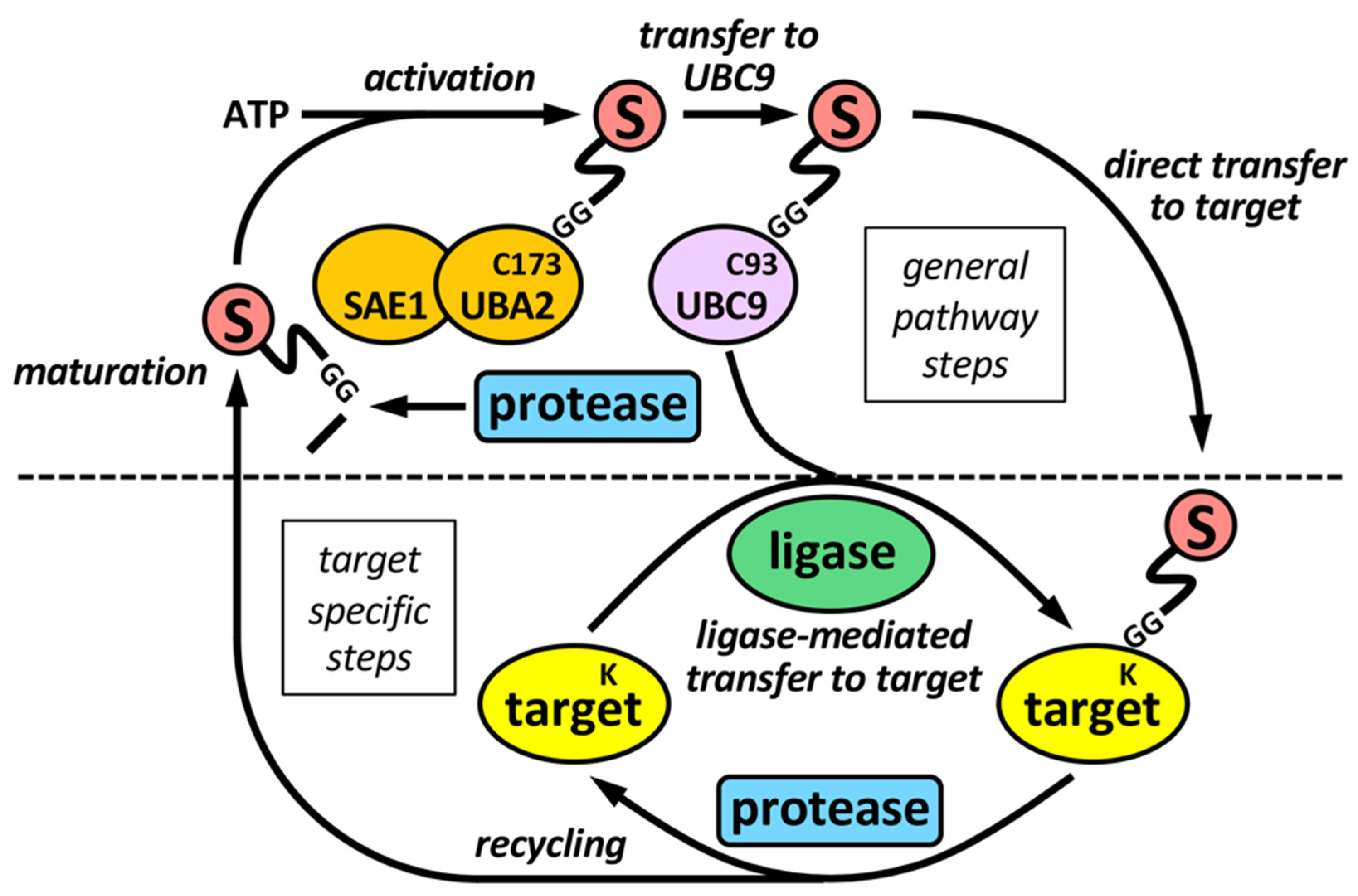 Figure 1. The sumoylation pathway. E1 enzyme (SAE1-UBA2 heterodimer), by using ATP, activates mature SUMO and transfers it to the E2 enzyme UBC9, which ultimately transfer SUMO to targets, either directly, or more frequently, assisted by an E3 SUMO ligase. SUMO maturation and scission from targets are performed by specific SUMO proteases. Maturation involves the exposition of two tandem Gly (G) residues at the C-terminus of mature SUMO. Activation and transfer to UBC9 involve the formation of thioester bonds between C-terminus of mature SUMO and specific Cys (C) residues at catalytic sites of E1 and E2, respectively. Defined Lys (K) residues at targets are the final SUMO acceptors for covalent attachment. Proteases and ligases are linked to target selection-associated steps in the sumoylation pathway.
Figure 1. The sumoylation pathway. E1 enzyme (SAE1-UBA2 heterodimer), by using ATP, activates mature SUMO and transfers it to the E2 enzyme UBC9, which ultimately transfer SUMO to targets, either directly, or more frequently, assisted by an E3 SUMO ligase. SUMO maturation and scission from targets are performed by specific SUMO proteases. Maturation involves the exposition of two tandem Gly (G) residues at the C-terminus of mature SUMO. Activation and transfer to UBC9 involve the formation of thioester bonds between C-terminus of mature SUMO and specific Cys (C) residues at catalytic sites of E1 and E2, respectively. Defined Lys (K) residues at targets are the final SUMO acceptors for covalent attachment. Proteases and ligases are linked to target selection-associated steps in the sumoylation pathway.2.1. SUMO Proteases
Proteases involved in SUMO maturation and recycling are SUMO specific, and the most studied are those of the SENP family (Figure 1 and Figure 2 and Table 1). This family includes SENP1–3 and SENP5–7 [21]. Besides, additional proteases also with specific activity on SUMO have been described more recently. They include a new family of desumoylating isopeptidases (DeSI) [22], which comprises DESI1 and DESI2, and USPL1 [23] and HINT1 [24] (Table 1). All SENP proteins display a desumoylation catalytic domain at the C-terminus, which appears split in the case of SENP6 and SENP7 (Figure 2A). Different sequences in the N-terminal region of SENPs have been related to cellular localization, and, in the case of SENP7, to interaction with the heterochromatin protein 1 (HP1) through two tandem PxVxL motifs [25]. Moreover, a SENP7 splice variant, lacking one of these motifs, has been reported to predict for good prognosis in breast cancer patients [26]. Additional variants have been described for other SENPs (https://www.ncbi.nlm.nih.gov/gene/?term=SENP (accessed on 14 July 2022)). SENP1 and SENP2 have been indicated to be able to maturate SUMO1–3, although SENP1 shows preference for SUMO1 and SENP2 for SUMO2 [21] (Figure 2B). SENP5 has been also shown to efficiently maturate SUMO2. Regarding SUMO recycling, SENP1 and SENP2 have been demonstrated to efficiently detach the three SUMO paralogs from targets, while SENP3 and SENP5 are more selective on SUMO2/3 and SENP6 and SENP7 display poly SUMO2/3 chains editing activity [21] (Figure 2B). In the cell, SENP1 and SENP2 have been associated with PML NBs in interphase (as SENP6) and with the nuclear pore complex (NPC); SENP3 and SENP5 with both the nucleolus and the mitochondria; and SENP6, SENP7 and SENP3 with chromatin [27] (Figure 2C).
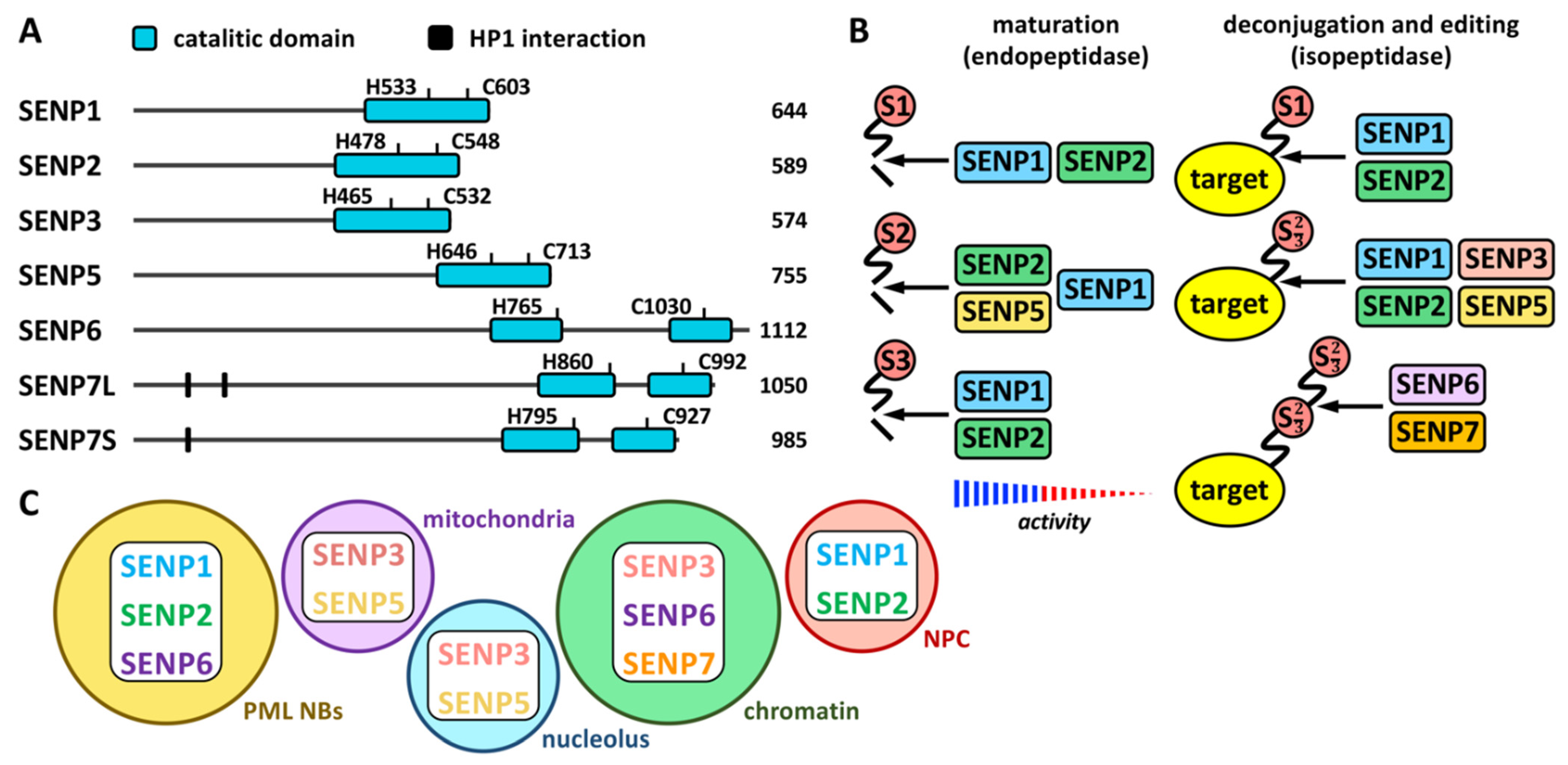 Figure 2. The SENP family. (A) Schematic representation of human SENP proteins with the C-terminal catalytic domain and the key His (H) and Cys (C) residues. HP1 interacting domains are also shown on SENP7, for which two isoforms have been described: large (SENP7L) and short (SENP7S), lacking the latter of one of the HP1 interacting motifs. The number of amino acids is also shown for each protein. (B) Endopeptidase (maturation) and isopeptidase (deconjugation and editing) activities for each SENP protein are shown. Strength of activity in relation to paralog preference is indicated for maturation activity. (C) Localization of SENP proteins to different cellular compartments is schematically represented. NPC, nuclear pore complex.
Figure 2. The SENP family. (A) Schematic representation of human SENP proteins with the C-terminal catalytic domain and the key His (H) and Cys (C) residues. HP1 interacting domains are also shown on SENP7, for which two isoforms have been described: large (SENP7L) and short (SENP7S), lacking the latter of one of the HP1 interacting motifs. The number of amino acids is also shown for each protein. (B) Endopeptidase (maturation) and isopeptidase (deconjugation and editing) activities for each SENP protein are shown. Strength of activity in relation to paralog preference is indicated for maturation activity. (C) Localization of SENP proteins to different cellular compartments is schematically represented. NPC, nuclear pore complex.2.2. SUMO Ligases
A key difference between SUMO proteases and ligases is that most SUMO ligases display additional functions independent of the SUMO ligase activity. This complicates in many cases the functional analysis in relation to the role of sumoylation. PIAS proteins (Figure 3), probably the most studied SUMO ligases, were among the first identified proteins displaying SUMO ligase activity, together with the NPC-associated protein RanBP2 and the Polycomb Repressive Complex 2 protein CBX4 (or PC2) [28]. Among these, PIASs and RanBP2 have been well characterized at the biochemical level (reviewed in [29]). The PIAS family comprises members 1–4. Two variants due to alternative splice have been classically described for PIAS2 (Figure 3A), although additional variants have been described for the different PIAS coding genes (https://www.ncbi.nlm.nih.gov/gene/?term=PIAS (accessed on 14 July 2022)). Structurally, PIAS proteins present an N-terminal SAP domain, involved in DNA binding as well as in interaction with transcriptional co-regulators, a PINIT region involved in nuclear localization, a RING-like SP-RING domain, an acidic stretch, and a C-terminal region rich in Ser/Thr (S/T) [30]. Different SIMs, relevant for PIAS function, have been described for the different members, both inside and outside the acidic stretch (reviewed in [31]). Most ligases are not required for in vitro modification of targets, which seems to depend exclusively on the presence of mature SUMO, E1 and E2. However, ligase requirement has been reported in vivo for a great variety of physiological processes. It was initially indicated that ligase activity in many cases relies in the ability of ligases to recruit at the same time, through different domains, SUMO-loaded UBC9 and the SUMO target to facilitate SUMO transfer, being the SP-RING domain the responsible for UBC9 recruitment in the case of PIAS proteins (Figure 3B). However, for RanBP2 ligase activity, target interaction seems to be dispensable, as minimal ligase appears to only require the simultaneous binding of SUMO1 and UBC9 to RanBP2 to optimally position the thioester bond for efficient transfer to its main target, RanGAP1 [32][33] (Figure 3B). Initial in vitro studies using the yeast PIAS1 ortholog Siz1 mapped the minimal E3 ligase domain to the region comprising the PINIT and the SP-RING motifs [34] (Figure 3B). Interestingly, another RING-related motif, the PHD domain, has been also implicated in UBC9 binding and sumoylation; for instance, in KAP1 and AtSIZ1 [35][36]. First studies on PIAS proteins also showed a significant level of promiscuity in target selection, although subsequent in vivo approaches have demonstrated more specific effects (reviewed in [18]). In contrast to ubiquitination that requires hundreds of E3 ligases for specific target selection, only a few dozens of ligases have been described for SUMO. Besides PIAS, RanBP2 and CBX4, enhanced sumoylation of specific targets has been associated with additional proteins, including: TOPORS, RSUME, MUL1, RHES, some proteins of the tripartite motif family (TRIM), ARF, SF2, class IIa histone deacetylases (HDACs), the PIAS-related proteins NSMCE2 and ZMIZ1-2, SLX4, KROX20, RNF212, UHRF2, TRAF7, ZBED1, MDM2 and ZNF451 (Table 1). Among TRIM proteins, SUMO ligase activity has been attributed to TRIM1, 11, 19, 22, 27, 28, 32, 33, 36, 38, 39 and L2 (Table 1). Interestingly, in some cases, as for TRIM27, and in relation to TP53, ligase activity has been reported for both SUMO and ubiquitin [37]. Additional proteins also show a dual function as E3 ubiquitin and SUMO ligases, such as UHRF2, TOPORS, TRAF7 or MDM2, and in some cases, distinct domains contribute to one or another activity [31]. Nevertheless, for some proteins dual function is controversial. For instance, TRIM25 has been described to stimulate TP53 sumoylation, but mechanistically this has been explained on the basis of TRIM25 requirement for recruitment of the RanBP2/G3BP2 complex, which ultimately mediates TP53 sumoylation [38]. Of note, ZNF451 has also been well characterized biochemically [39], and it has been proposed to display elongase E4 activity besides E3 ligase activity, due to its ability to assemble poly-SUMO2/3 chains through the action of tandem SIMs at the N-terminus [40]. Another open reading frame (ORF): KIAA 1586, close to ZNF451, encodes a protein with similar N-terminal tandem SIMs, which has also proved to display elongase activity. Known SUMO ligases are quite divergent phylogenetically and structurally and, as indicated, there are not defined families of proteins exclusively devoted to this function, which seems to appear in a high variety of proteins with additional functions. Thus, it seems likely that well-known proteins will reveal unexpected SUMO ligase activity in the near future.
Table 1. SUMO proteases and ligases.
| Enzymes | Family | Proteins | References |
|---|---|---|---|
| Proteases | SENP | SENP1, SENP2, SENP3, SENP5, SENP6, SENP7 |
[21] |
| USPL1 | USPL1 | [23] | |
| DeSI | DESI1, DESI2 | [22] | |
| Ligases | SP-RING | PIAS1, PIAS2, PIAS3, PIAS4 | [28][30][31] |
| NSMCE2 (NSE2/MMS21) | |||
| ZMIZ1 (ZIMP10), ZMIZ2 | |||
| TRIM | TRIM1, TRIM11, TRIM19 (PML), TRIM22, TRIM27, TRIM28 (KAP1), TRIM32, TRIM33 (TIF1g), TRIM36, TRIM38, TRIM39, TRIML2 | [37][41][42][43][44][45] | |
| HDACs IIa | HDAC4, HDAC5, HDAC7, HDAC9 (MITR) |
[46] | |
| Elongases | ZNF451 | [39][40] | |
| KIAA 1586 | |||
| Others | RanBP2 | [18][28][31][47][48][49] | |
| CBX4 (PC2) | |||
| TOPORS | |||
| RSUME | |||
| MUL1 (MAPL) | |||
| RHES | |||
| ARF (P14) | |||
| SF2 (ASF) | |||
| SLX4 | |||
| KROX20 | |||
| RNF212 | |||
| UHRF2 (RNF107) | |||
| TRAF7 (RNF119) | |||
| BCA2 (RNF115) | |||
| ZBED1 (DREF) | |||
| MDM2 |
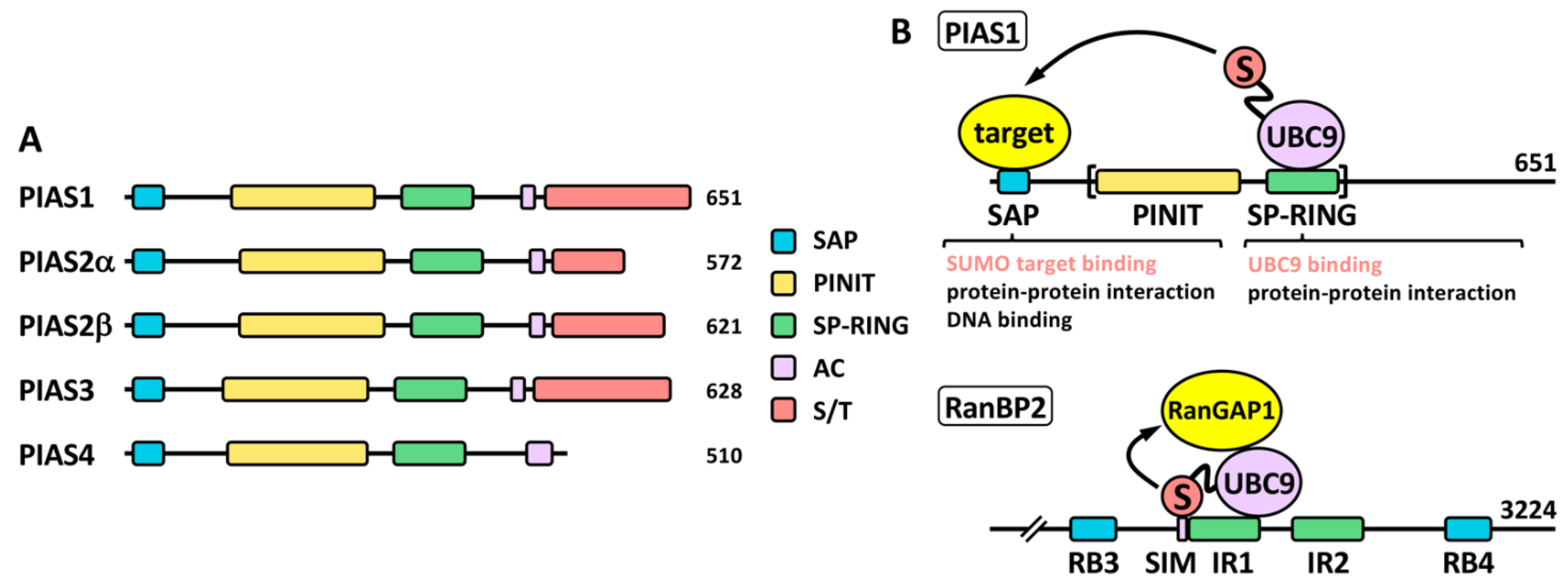 Figure 3. The PIAS family. (A) Schematic representation of human PIAS proteins. The SP-RING and SAP domains, together with the PINIT region, the acidic stretch (AC) and the C-terminal Ser/Thr (S/T) rich region, are shown. Two alternative isoforms, α and β, have been classically described for PIAS2. The number of amino acids is also shown for each protein. (B) Proposed ligase mechanisms using PIAS1 and RanBP2 as models. In the case of PIAS1, simultaneous binding of target and SUMO-loaded UBC9 through different domains may facilitate SUMO transfer to the target. The SP-RING is important for UBC9 binding but may also mediate other protein interactions for additional functions. Other domains, like the SAP domain, may recruit SUMO targets, but it is also involved in additional functions. The minimal protein region required for ligase activity is indicated with brackets. In the case of RanBP2, interaction of SUMO and UBC9 with a SIM and the 50-amino acid internal repeat (IR) 1, respectively, should position the thioester bond in the appropriate conformation for efficient transfer of SUMO to the target, which is not required to directly interact with the ligase. RB, Ran binding domain.
Figure 3. The PIAS family. (A) Schematic representation of human PIAS proteins. The SP-RING and SAP domains, together with the PINIT region, the acidic stretch (AC) and the C-terminal Ser/Thr (S/T) rich region, are shown. Two alternative isoforms, α and β, have been classically described for PIAS2. The number of amino acids is also shown for each protein. (B) Proposed ligase mechanisms using PIAS1 and RanBP2 as models. In the case of PIAS1, simultaneous binding of target and SUMO-loaded UBC9 through different domains may facilitate SUMO transfer to the target. The SP-RING is important for UBC9 binding but may also mediate other protein interactions for additional functions. Other domains, like the SAP domain, may recruit SUMO targets, but it is also involved in additional functions. The minimal protein region required for ligase activity is indicated with brackets. In the case of RanBP2, interaction of SUMO and UBC9 with a SIM and the 50-amino acid internal repeat (IR) 1, respectively, should position the thioester bond in the appropriate conformation for efficient transfer of SUMO to the target, which is not required to directly interact with the ligase. RB, Ran binding domain.Pathway components can be influenced by the physiological environment, which may have an impact in their activities. In this sense, several SENPs have been shown to be sensible to hypoxic inactivation [50]. In addition, SENP3 has been defined as a redox sensor, and, for instance, reactive oxygen species (ROS) may modulate E1 activity (reviewed in [51]). Besides, a number of PTMs, including phosphorylation, acetylation, ubiquitination and sumoylation itself, have been shown to affect several pathway components. Otherwise, a patent crosstalk between sumoylation and other PTMs like ubiquitination, phosphorylation and methylation operates on target modification [4][28][52]. As indicated, vertebrate viability depends on sumoylation, and acting on E1 or E2, which will globally affect sumoylation, should certainly have general and strong effects. However, the function of regulating modification of specific targets mostly relies on SUMO proteases and ligases. Target modification output depends on localization, expression, activity and paralog preference of these enzymes. Thus, their unbalanced expression, localization or function, directly impact on sumoylation of defined proteins, which in many cases is in the basis of physiological alterations leading to tumorigenesis. Depending on the specific association of particular ligases and proteases with precise targets, and, thereby, on the nature of these and the affected tissue, alterations will lead to the appearance of a great variety of cancer types.
References
- Sahin, U.; de The, H.; Lallemand-Breitenbach, V. Sumoylation in Physiology, Pathology and Therapy. Cells 2022, 11, 814.
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140.
- Hendriks, I.A.; Lyon, D.; Su, D.; Skotte, N.H.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018, 9, 2456.
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.; Nielsen, M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336.
- Hendriks, I.A.; Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595.
- Bernstock, J.D.; Yang, W.; Ye, D.G.; Shen, Y.; Pluchino, S.; Lee, Y.J.; Hallenbeck, J.M.; Paschen, W. SUMOylation in brain ischemia: Patterns, targets, and translational implications. J. Cereb. Blood Flow Metab. 2018, 38, 5–16.
- Enserink, J.M. Sumo and the cellular stress response. Cell Div. 2015, 10, 4.
- Niskanen, E.A.; Palvimo, J.J. Chromatin SUMOylation in heat stress: To protect, pause and organise?: SUMO stress response on chromatin. Bioessays 2017, 39, 1600263.
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 1997, 88, 97–107.
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470.
- Lascorz, J.; Codina-Fabra, J.; Reverter, D.; Torres-Rosell, J. SUMO-SIM interactions: From structure to biological functions. Semin. Cell Dev. Biol. 2021, S1084-9521(21)00283-4.
- Yau, T.Y.; Sander, W.; Eidson, C.; Courey, A.J. SUMO Interacting Motifs: Structure and Function. Cells 2021, 10, 2825.
- Sriramachandran, A.M.; Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 2014, 1843, 75–85.
- Sahin, U.; Ferhi, O.; Jeanne, M.; Benhenda, S.; Berthier, C.; Jollivet, F.; Niwa-Kawakita, M.; Faklaris, O.; Setterblad, N.; de The, H.; et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014, 204, 931–945.
- Jentsch, S.; Psakhye, I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 2013, 47, 167–186.
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12.
- Nacerddine, K.; Lehembre, F.; Bhaumik, M.; Artus, J.; Cohen-Tannoudji, M.; Babinet, C.; Pandolfi, P.P.; Dejean, A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 2005, 9, 769–779.
- Garcia-Gutierrez, P.; Garcia-Dominguez, M. SUMO control of nervous system development. Semin. Cell Dev. Biol. 2021, S1084-9521(21)00304-9.
- Wang, L.; Wansleeben, C.; Zhao, S.; Miao, P.; Paschen, W.; Yang, W. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014, 15, 878–885.
- Chymkowitch, P.; Nguea, P.A.; Enserink, J.M. SUMO-regulated transcription: Challenging the dogma. Bioessays 2015, 37, 1095–1105.
- Kunz, K.; Piller, T.; Muller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131, jcs211904.
- Shin, E.J.; Shin, H.M.; Nam, E.; Kim, W.S.; Kim, J.H.; Oh, B.H.; Yun, Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012, 13, 339–346.
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938.
- Cortes-Montero, E.; Rodriguez-Munoz, M.; Sanchez-Blazquez, P.; Garzon, J. The Axonal Motor Neuropathy-Related HINT1 Protein Is a Zinc- and Calmodulin-Regulated Cysteine SUMO Protease. Antioxid. Redox Signal. 2019, 31, 503–520.
- Romeo, K.; Louault, Y.; Cantaloube, S.; Loiodice, I.; Almouzni, G.; Quivy, J.P. The SENP7 SUMO-Protease Presents a Module of Two HP1 Interaction Motifs that Locks HP1 Protein at Pericentric Heterochromatin. Cell Rep. 2015, 10, 771–782.
- Bawa-Khalfe, T.; Lu, L.S.; Zuo, Y.; Huang, C.; Dere, R.; Lin, F.M.; Yeh, E.T. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA 2012, 109, 17466–17471.
- Huang, C.J.; Wu, D.; Khan, F.A.; Huo, L.J. DeSUMOylation: An Important Therapeutic Target and Protein Regulatory Event. DNA Cell Biol. 2015, 34, 652–660.
- Salas-Lloret, D.; Gonzalez-Prieto, R. Insights in Post-Translational Modifications: Ubiquitin and SUMO. Int. J. Mol. Sci. 2022, 23, 3281.
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385.
- Rabellino, A.; Andreani, C.; Scaglioni, P.P. The Role of PIAS SUMO E3-Ligases in Cancer. Cancer Res. 2017, 77, 1542–1547.
- Shi, X.; Du, Y.; Li, S.; Wu, H. The Role of SUMO E3 Ligases in Signaling Pathway of Cancer Cells. Int. J. Mol. Sci. 2022, 23, 3639.
- Pichler, A.; Knipscheer, P.; Saitoh, H.; Sixma, T.K.; Melchior, F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat. Struct. Mol. Biol. 2004, 11, 984–991.
- Reverter, D.; Lima, C.D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 2005, 435, 687–692.
- Takahashi, Y.; Kikuchi, Y. Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J. Biol. Chem. 2005, 280, 35822–35828.
- Garcia-Dominguez, M.; March-Diaz, R.; Reyes, J.C. The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. 2008, 283, 21469–21477.
- Ivanov, A.V.; Peng, H.; Yurchenko, V.; Yap, K.L.; Negorev, D.G.; Schultz, D.C.; Psulkowski, E.; Fredericks, W.J.; White, D.E.; Maul, G.G.; et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 2007, 28, 823–837.
- Chu, Y.; Yang, X. SUMO E3 ligase activity of TRIM proteins. Oncogene 2011, 30, 1108–1116.
- Takayama, K.I.; Suzuki, T.; Tanaka, T.; Fujimura, T.; Takahashi, S.; Urano, T.; Ikeda, K.; Inoue, S. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene 2018, 37, 2165–2180.
- Cappadocia, L.; Pichler, A.; Lima, C.D. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 2015, 22, 968–975.
- Eisenhardt, N.; Chaugule, V.K.; Koidl, S.; Droescher, M.; Dogan, E.; Rettich, J.; Sutinen, P.; Imanishi, S.Y.; Hofmann, K.; Palvimo, J.J.; et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 2015, 22, 959–967.
- Hannoun, Z.; Maarifi, G.; Chelbi-Alix, M.K. The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev. 2016, 29, 3–16.
- Hu, M.M.; Yang, Q.; Xie, X.Q.; Liao, C.Y.; Lin, H.; Liu, T.T.; Yin, L.; Shu, H.B. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569.
- Ikeuchi, Y.; Dadakhujaev, S.; Chandhoke, A.S.; Huynh, M.A.; Oldenborg, A.; Ikeuchi, M.; Deng, L.; Bennett, E.J.; Harper, J.W.; Bonni, A.; et al. TIF1gamma protein regulates epithelial-mesenchymal transition by operating as a small ubiquitin-like modifier (SUMO) E3 ligase for the transcriptional regulator SnoN1. J. Biol. Chem. 2014, 289, 25067–25078.
- Kung, C.P.; Khaku, S.; Jennis, M.; Zhou, Y.; Murphy, M.E. Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of proapoptotic genes. Mol. Cancer Res. 2015, 13, 250–262.
- Zhu, G.; Harischandra, D.S.; Ghaisas, S.; Zhang, P.; Prall, W.; Huang, L.; Maghames, C.; Guo, L.; Luna, E.; Mack, K.L.; et al. TRIM11 Prevents and Reverses Protein Aggregation and Rescues a Mouse Model of Parkinson’s Disease. Cell Rep. 2020, 33, 108418.
- Gregoire, S.; Yang, X.J. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 2005, 25, 2273–2287.
- Shi, Y.; Castro-Gonzalez, S.; Chen, Y.; Serra-Moreno, R. Effects of the SUMO Ligase BCA2 on Metabolic Activity, Cell Proliferation, Cell Migration, Cell Cycle, and the Regulation of NF-kappaB and IRF1 in Different Breast Epithelial Cellular Contexts. Front. Cell Dev. Biol. 2021, 9, 711481.
- Stindt, M.H.; Carter, S.; Vigneron, A.M.; Ryan, K.M.; Vousden, K.H. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle 2011, 10, 3176–3188.
- Talamillo, A.; Barroso-Gomila, O.; Giordano, I.; Ajuria, L.; Grillo, M.; Mayor, U.; Barrio, R. The role of SUMOylation during development. Biochem. Soc. Trans. 2020, 48, 463–478.
- Kunz, K.; Wagner, K.; Mendler, L.; Holper, S.; Dehne, N.; Muller, S. SUMO Signaling by Hypoxic Inactivation of SUMO-Specific Isopeptidases. Cell Rep. 2016, 16, 3075–3086.
- Henley, J.M.; Craig, T.J.; Wilkinson, K.A. Neuronal SUMOylation: Mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014, 94, 1249–1285.
- Stabell, M.; Saether, T.; Rohr, A.K.; Gabrielsen, O.S.; Myklebost, O. Methylation-dependent SUMOylation of the architectural transcription factor HMGA2. Biochem. Biophys. Res. Commun. 2021, 552, 91–97.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Revisions:
2 times
(View History)
Update Date:
25 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





