Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Patrick Caffrey | -- | 4545 | 2022-07-22 15:33:54 | | | |
| 2 | Amina Yu | -76 word(s) | 4469 | 2022-07-25 03:50:15 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Caffrey, P.; Hogan, M.; Song, Y. New Glycosylated Polyene Macrolides. Encyclopedia. Available online: https://encyclopedia.pub/entry/25443 (accessed on 04 March 2026).
Caffrey P, Hogan M, Song Y. New Glycosylated Polyene Macrolides. Encyclopedia. Available at: https://encyclopedia.pub/entry/25443. Accessed March 04, 2026.
Caffrey, Patrick, Mark Hogan, Yuhao Song. "New Glycosylated Polyene Macrolides" Encyclopedia, https://encyclopedia.pub/entry/25443 (accessed March 04, 2026).
Caffrey, P., Hogan, M., & Song, Y. (2022, July 22). New Glycosylated Polyene Macrolides. In Encyclopedia. https://encyclopedia.pub/entry/25443
Caffrey, Patrick, et al. "New Glycosylated Polyene Macrolides." Encyclopedia. Web. 22 July, 2022.
Copy Citation
Polyene macrolides are antifungal agents that are synthesized by actinomycetes and other bacteria. These compounds consist of macrolactone rings containing between three and eight conjugated double bonds. Most are glycosylated with a single aminodeoxysugar and specifically bind ergosterol in fungal cell membranes.
glycosylated polyene macrolide
antifungal antibiotics
biosynthetic gene clusters
1. Characteristic Features of Polyene Biosynthetic Gene Clusters (BGCs)
The macrolactone rings of glycosylated polyene macrolides (GPMs) are synthesized by modular polyketide synthases. There is now a detailed understanding of how these enzymatic assembly lines select starter and extender units, determine alcohol and methyl stereochemistry, and impose double-bond geometry [1]. Within the multienzyme polypeptides, acyltransferase (AT), ketoreductase (KR), and enoylreductase (ER) domains contain conserved motifs that correlate with the stereochemical outcome of each reaction catalyzed. In some cases, structural studies have given insights into the functions of the key amino acids in the reaction mechanism. Bioinformatic analysis now allows accurate prediction of polyketide stereostructures from amino acid sequences of PKS enzymes [1][2][3][4][5][6]. These prediction methods are a useful aid to chemical methods for determination of polyketide structures in general. Stereostructures predicted from silent polyene PKSs maintain the stereochemical homology found in experimentally characterized GPMs (Figure 1) [7][8].
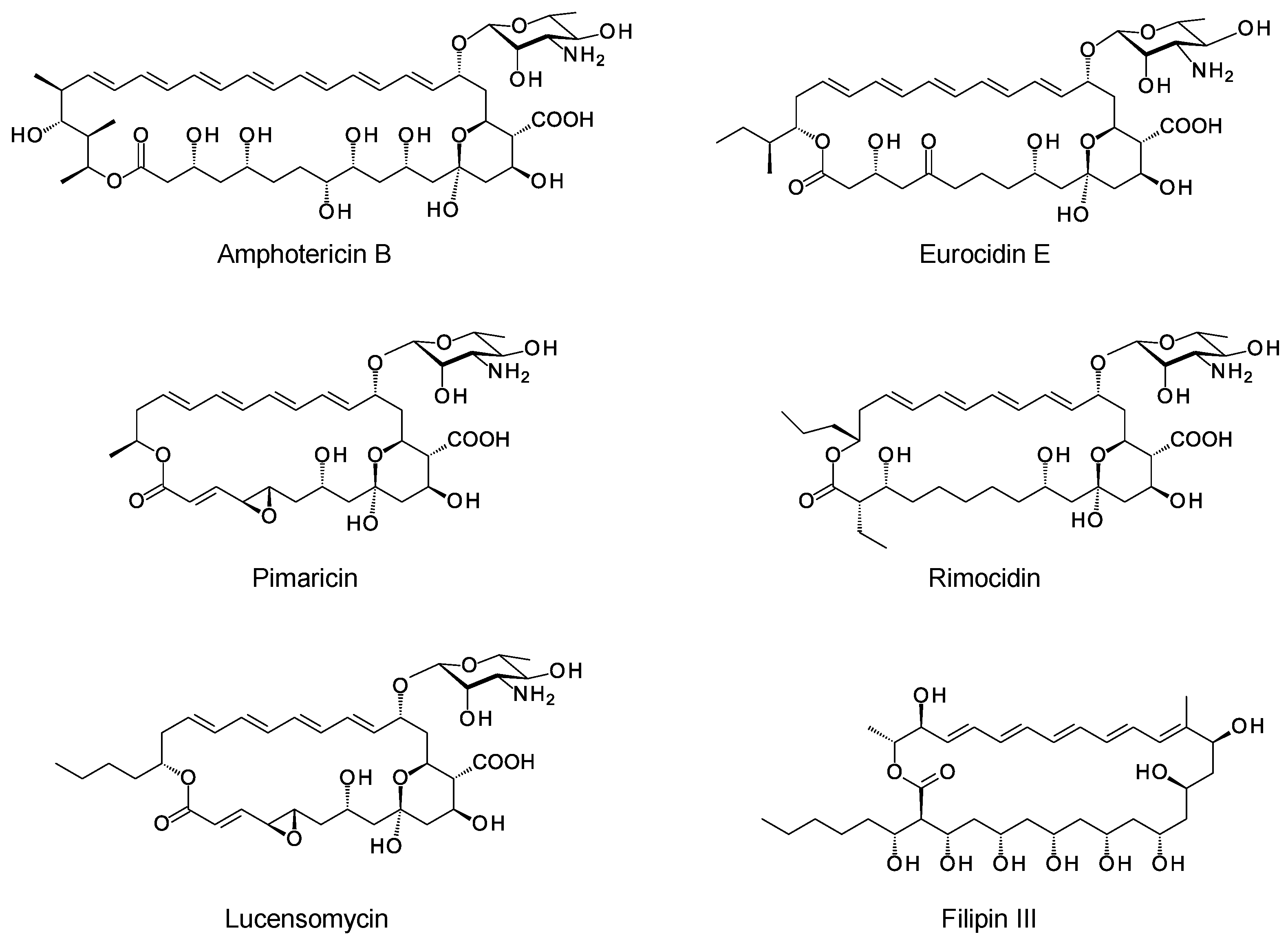
Figure 1. Structures of polyene macrolides.
A typical polyene PKS has a series of modules for synthesis of a polyene unit containing at least four conjugated double bonds [9]. This is followed by a hexamodular PKS protein capable of synthesizing a region that forms a hemiketal, includes a methyl branch that is oxidized to a carboxyl group, and a hydroxyl group that is glycosylated with mycosamine (Figure 2) [9]. In the amphotericin PKS, this hexamodular protein is AmphI. The cluster contains genes for a cytochrome P450 (AmphN) that oxidizes the methyl branch, GDP-α-d-mycosamine synthase (AmphDII), and a d-mycosamine-specific glycosyltransferase (AmphDI). A GDP-α-d-mannose 4,6-dehydratase (AmphDIII) functions at an early stage in mycosamine biosynthesis [10]. Homologues of these genes are typical of polyene BGCs. BlastP searches with AmphI, AmphN, and AmphDI can reveal new clusters [11][12]. Additional P450s may modify the polyol chain further [13][14]. These modifications occur at different positions in different polyenes, so it is not possible to predict the precise functions of homologues of these P450s in silent clusters.
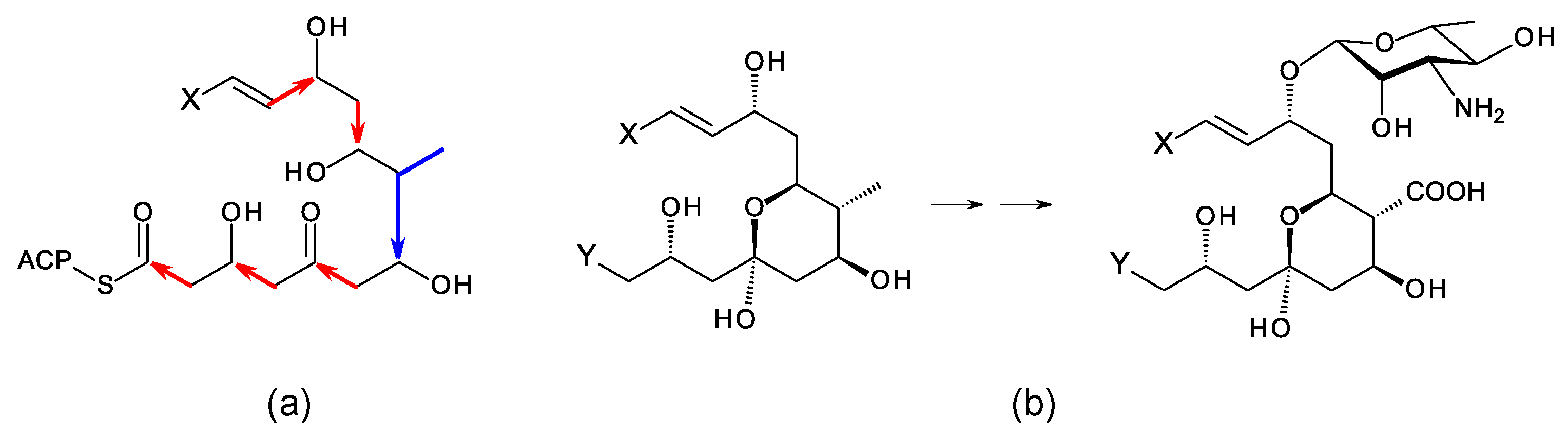
Figure 2. Conserved hemiketal structure that forms in polyene macrolides. (a) A hexamodular polyketide synthase protein synthesizes this region of the polyketide chain from malonyl-derived acetate (red arrows) and methylmalonyl-derived propionate (blue arrow) building blocks. (b) Exocyclic carboxyl group formation and glycosylation with mycosamine occur in this region. X represents the primer end of the polyketide chain, and Y represents the carboxy-terminal end.
There are a few examples of polyenes that lack exocyclic carboxyl groups or are glycosylated with sugars other than d-mycosamine. These are mentioned later in the text. d-mycosamine is not unique to GPMs but also appears in vancoresmycin, a non-polyene macrolide that disrupts membranes of Gram-positive bacteria [15][16]. The vancoresmycin BGC also contains homologues of AmphDIII, AmphDII, and AmphDI.
2. Silent Polyene PKSs Predicted to Use a Wider Range of Starter and Extender Units
For the well-studied polyene antibiotics, polyketide biosynthesis is primed with a limited range of starter units. Acetyl primers are used in the biosynthesis of amphotericin, nystatin, and pimaricin; a propionyl primer is used for selvamicin and lucensomycin; a butyryl primer is used for rimocidin; and p-aminobenzoate is used for aromatic heptaenes [7][17][18]. An analysis of the recently sequenced Streptomyces eurocidicus genome indicates that the eurocidin PKS uses (2S)-2-methylbutyrate as a primer [12]. An analysis of BGCs in genome sequences indicates that primers such as guanidinobutyrate, 3-amino-5-hydroxy-benzoic acid, and 3-hydroxybenzoic acid may be used by some polyene PKSs. Some of these PKSs are predicted to use ethylmalonyl and methoxymalonyl CoA extender units, which diversify macrolactone structures further. This section describes some specific examples. Genome accession numbers and isolation sources of microorganisms discussed are given. The supplementary figures contain more detail on how polyene structures were predicted.
2.1. Guanidinobutyrate Primers
A few polyketides are known for which biosynthesis of the polyketide chain begins with arginine-derived guanidinobutyryl or 4-aminobutyryl primers (Figure 3). These include antifungal linear polyene polyols ECO-02301 from Streptomyces aizunensis NRRL B-11277 [19], clethramycin from Streptomyces malaysiensis DSM4137, and mediomycin from Streptomyces mediocidicus [20][21][22] (Figure 3). Three enzymes convert l-arginine to guanidinobutyryl CoA: a decarboxylating arginine mono-oxygenase, an amide hydrolase, and a CoA ligase [20] (Figure 4). The ECO-020301 cluster also contains a gene for an amidinohydrolase that cleaves the guanidinobutyryl CoA so that a 4-aminobutyryl chain migrates onto the loading ACP to initiate polyketide biosynthesis (Figure 3 and Figure 4). An intact guanidinobutyryl unit acts as a primer for biosynthesis of clethramycin and mediomycin (Figure 4). In S. malaysiensis, the guanidinobutyryl primer remains in the final polyene, clethramycin. S. mediocidicus initially synthesizes the same polyene, but the guanidino group is hydrolyzed as a late modification, releasing urea and leaving a 4-aminobutyryl moiety as the remainder from the starter unit. The final product is mediomycin A1. The amidinohydrolase involved (Medi4948) is encoded by a gene located 670 kb distant from the mediomycin BGC in the S. mediocidicus chromosome [21].
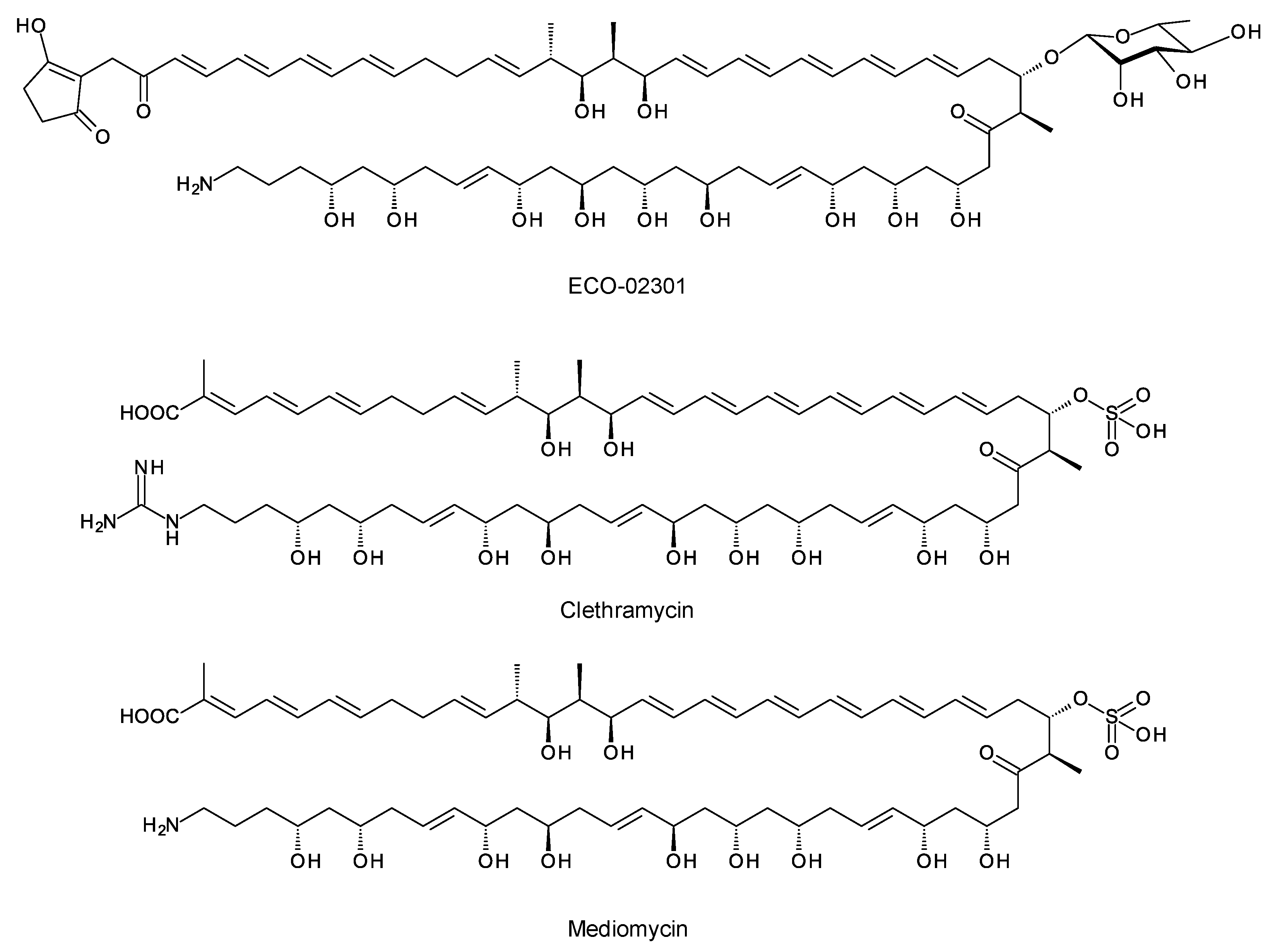
Figure 3. Structures of ECO-02031, clethramycin, and mediomycin.
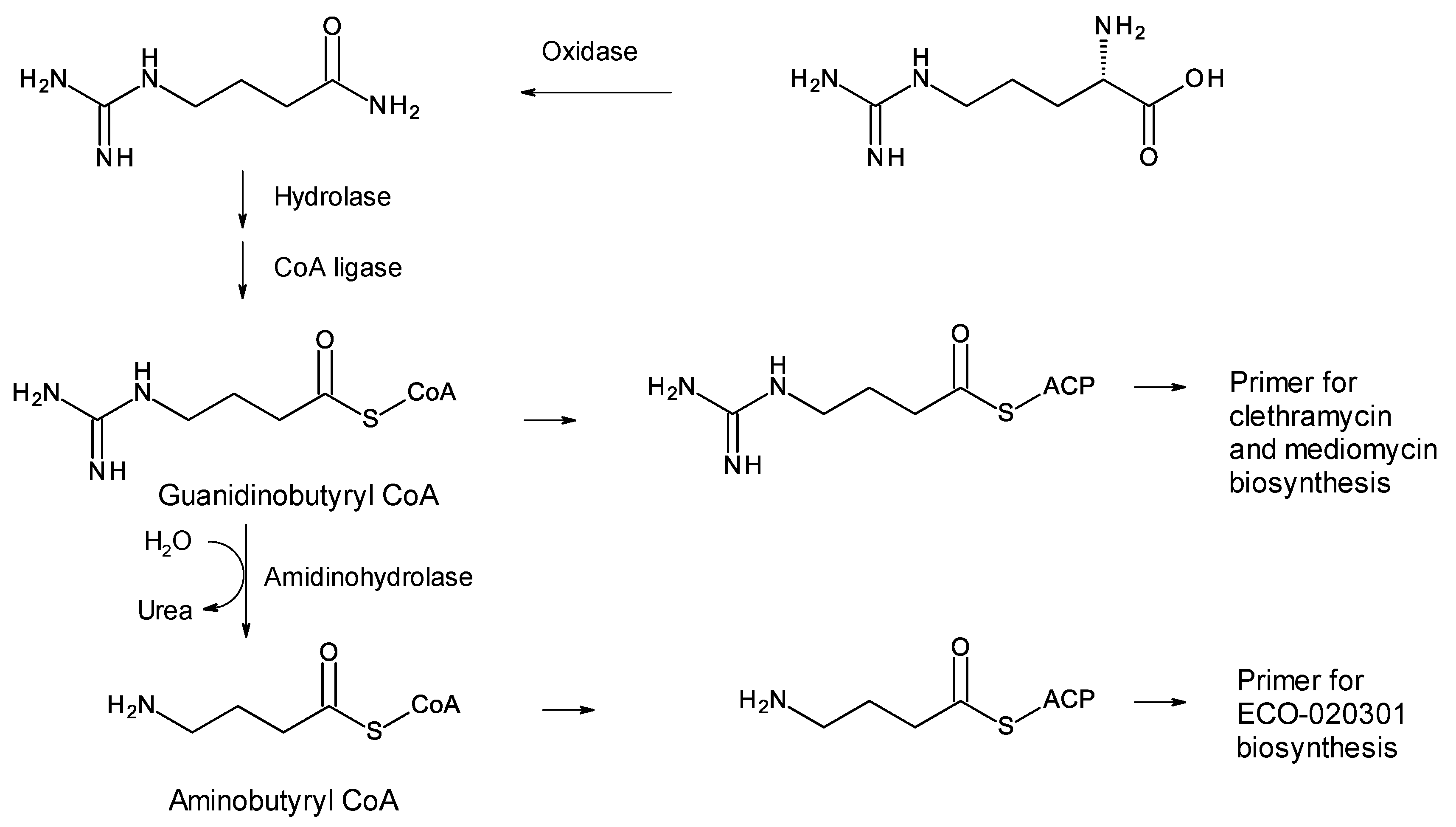
Figure 4. Formation of primers for biosynthesis of linear polyene polyols ECO-020301, clethramycin, and mediomycin.
Amycolatopsis saalfeldensis has a polyene BGC that includes homologues (57% identical and 71% similar) of the amine oxidase and acyl CoA synthetase that generate guanidinobutyryl CoA as a primer. The PKS assembly line starts with a loading ACP homologous to that for azalomycin PKS, which also uses a guanidinobutyryl primer [22]. The predicted polyene structure is shown in Figure 5. It is not possible to predict whether the guanidinobutyryl starter is cleaved after incorporation. The purification and characterization of these polyenes would be necessary to determine the structures correctly. Related BGCs are present in the genomes of Amycolatopsis jejuensis and Amycolatopsis benzoatilytica, but these are incompletely sequenced.
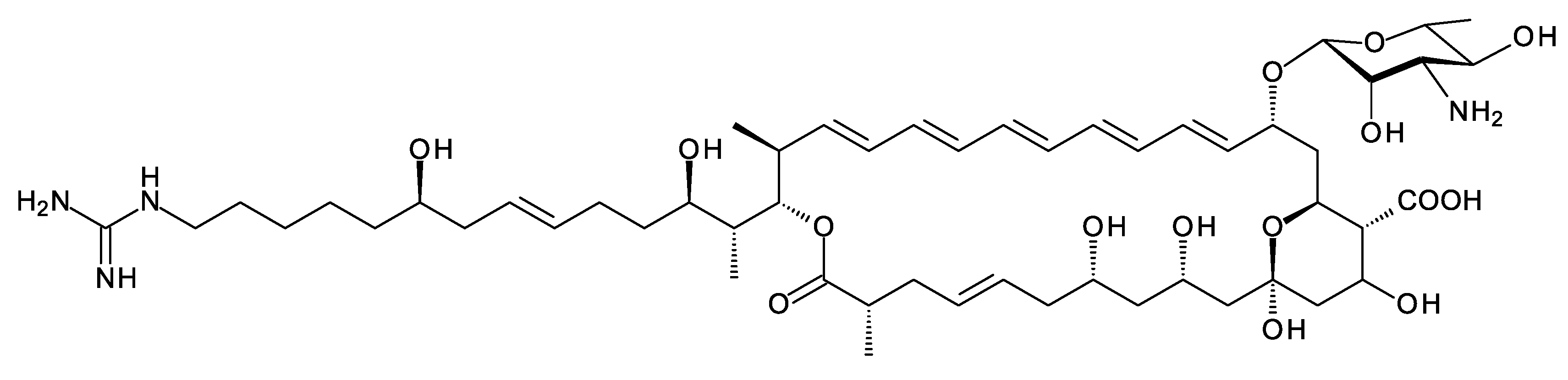
Figure 5. Predicted partial structure of the A. saalfeldensis polyene. The structure-prediction method used is given in the 2017 paper by Sheehan et al. [23].
So far, it has not been possible to detect the production of these polyenes in cultures of Amyc. saalfeldensis and Amyc. benzoatilytica [24]. These pentaenes are of interest because the presence of an amino or guanidino group may increase antifungal activity. In general, a net-positive charge increases the antimicrobial activity of a biocide that acts on membranes [25]. This may result from initial electrostatic interactions between the negatively charged microbial cell surfaces and positively charged surfactants.
2.2. Primer: 3-Amino-5-hydroxybenzoate
AHBA, 3-Amino-5-hydroxybenzoic acid, is used in the biosynthesis of several natural products. It serves as a primer for biosynthesis of ansamycin polyketides such as rifamycin and chaxamycin [26][27][28]. In the producers of these polyketides, AHBA is synthesized from kanosamine 6-phosphate in a complex pathway that requires seven enzymes [26].
Amycolatopsis albispora was found in a deep-sea sediment sample taken from the Indian Ocean [29]. The genome has a polyene BGC that includes genes for biosynthesis of AHBA from kanosamine [26], and the PKS loading module starts with a CoA ligase domain that is predicted to activate AHBA. AntiSMASH analysis predicts that extension modules 6 and 8 use ethylmalonyl CoA as extender unit but genes for biosynthesis of this extender are absent from the cluster. The structure prediction for the Amyc. albispora heptaene is shown in Figure 6. The extra amino group might increase activity. Amycolatopsis YIM10 has a similar cluster that is incompletely sequenced [30].
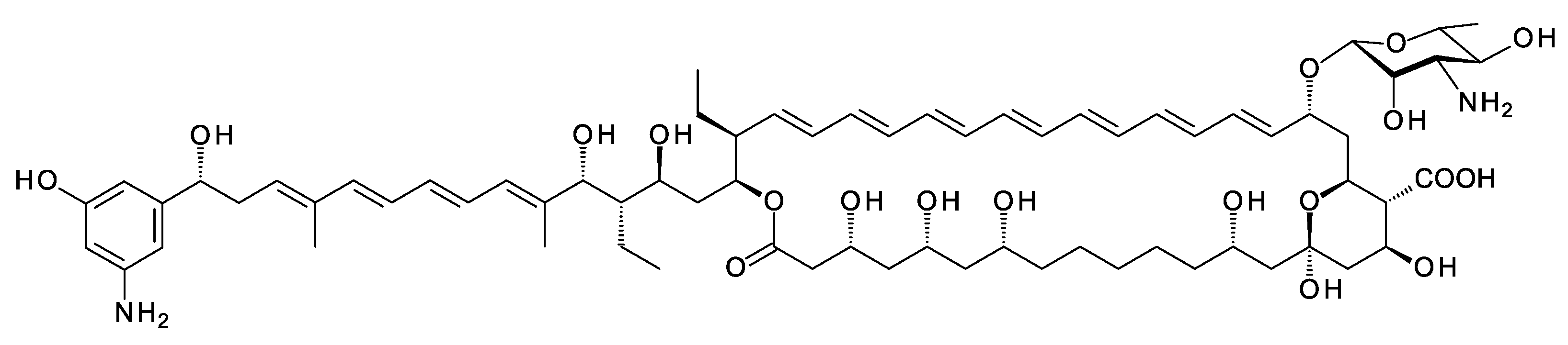
Figure 6. Predicted partial structure of Amyc. albispora heptaene. The BGC contains a gene for an AmphL cytochrome P450 homologue that may hydroxylate the polyol chain between C2 and C14. No prediction is made for this modification.
2.3. Starter: 3-Hydroxybenzoate
Two Saccharopolyspora species and two Lentzea species are predicted to synthesize similar heptaenes that are primed with 3-hydroxybenzoyl units. The Saccharopolyspora dendrathemae genome has a complete BGC for an all trans heptaene. A similar BGC in the genome of Saccharopolyspora flava has been described by Usachova [31]. The sequence of this cluster has not been disclosed but an independently determined incomplete version of the genome sequence for Sacc. flava DSM 44771 is publicly available (accession number = NZ_FOZX01000003). The PKSs start with CoA ligase domains that are predicted to activate a substituted benzoic acid as primer for polyketide chain initiation. Similar incomplete polyene PKS assembly lines occur in Lentzea xinjiangensis and Lentzea waywayandensis DSM44232. All four microorganisms lack genes for AHBA biosynthesis but have a gene for Hyg5 chorismatase that converts 9horismite to 3-hydroxybenzoate (Figure 7) [31][32][33].
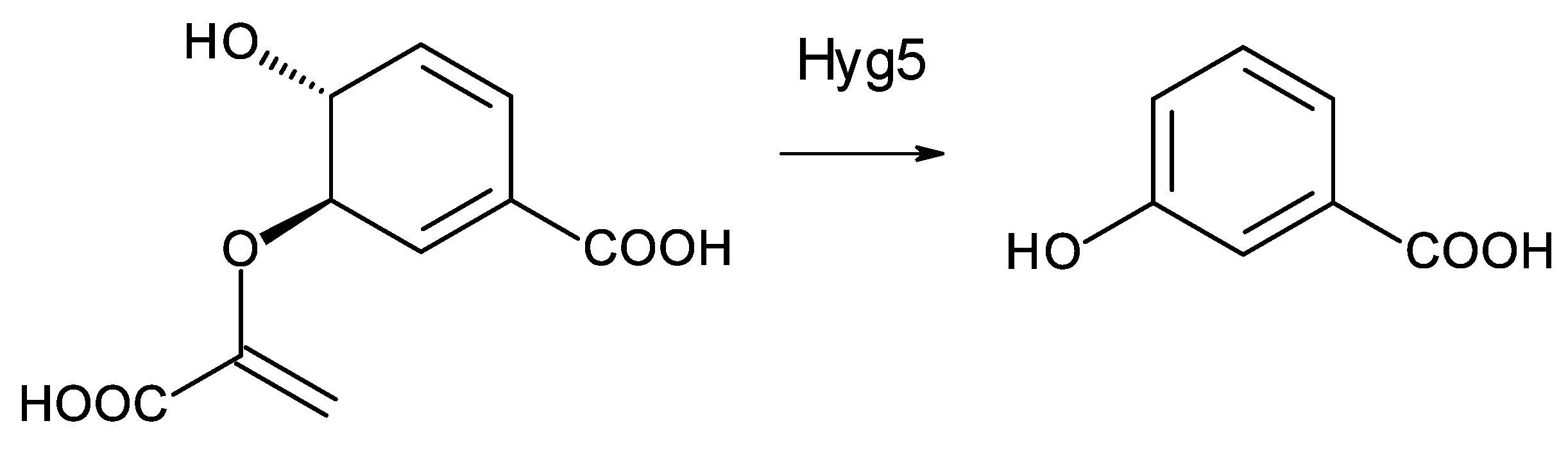
Figure 7. Conversion of chorismate to 3-hydroxybenzoate catalyzed by Hyg5. Homologues of Hyg5 occur in the polyene BGCs of Sacc. flava (WP_093416095.1), Sacc. dendrathemae (WP_145741796.1), Lentz. Waywayandensis (WP_093588251), and Lentz. Xingjiangensis (WP_089951167.1).
The Leadlay group overproduced the Sacc. flava chorismatase and reconstituted 3-hydroxybenzoate formation in vitro [31]. The Sacc. dendrathemae and Sacc. flava clusters both encode glycosyltransferases (GTs) homologous to AmphDI (62% identity), but there are no GDP-mannose DH or mycosamine synthase genes in the genomes of either microorganism. AmphDI is capable of using GDP-α-d-mannose as an alternative activated sugar donor [34], so these polyenes are possibly glycosylated with d-mannose. Alternatively, they may be synthesized as aglycones. Predicted structures of the two heptaenes from Sacc. dendrathemae and Sacc. flava are shown in Figure 8.
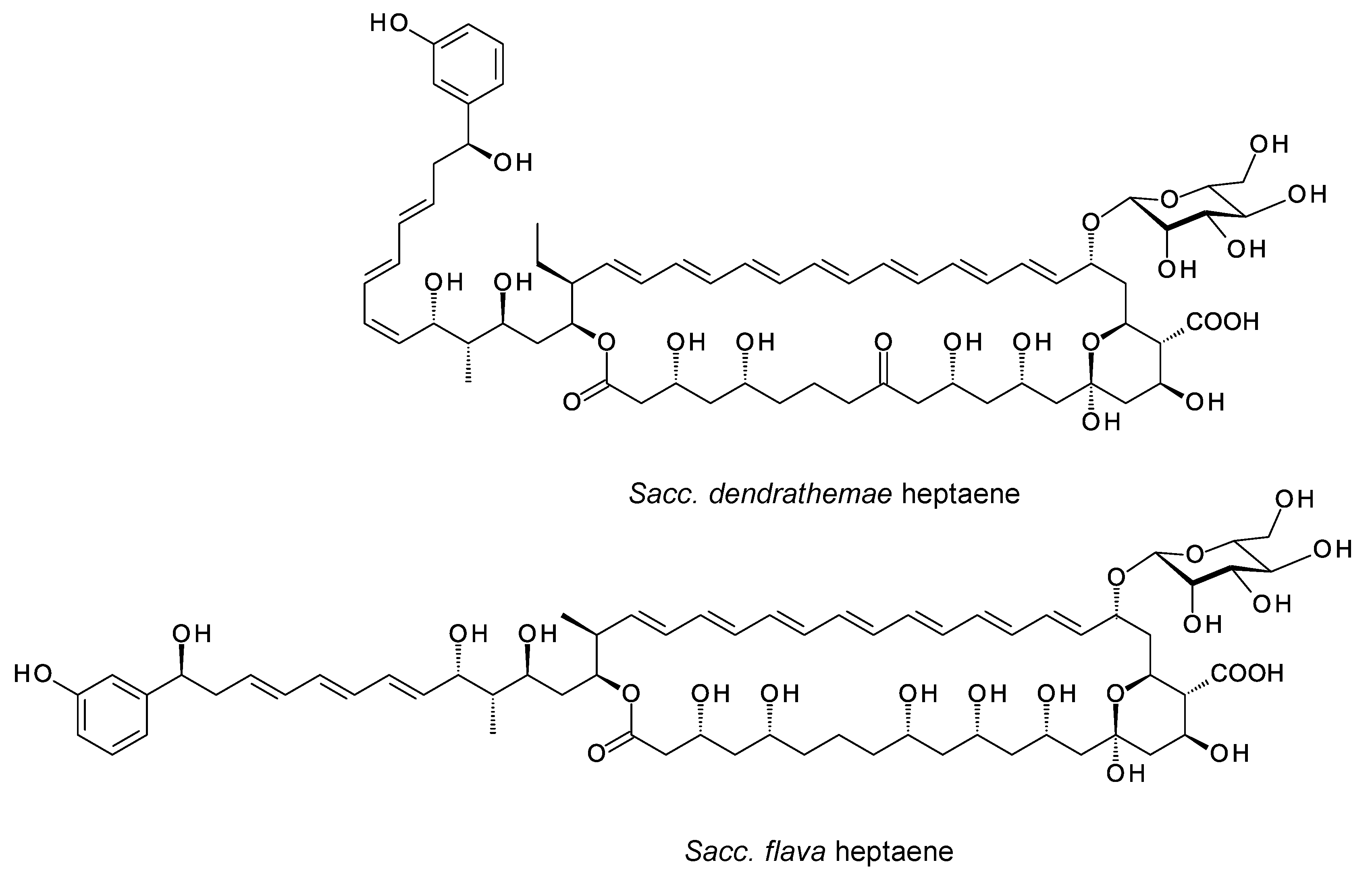
Figure 8. Predicted partial structures of heptaenes made by Sacc. dendrathemae and Sacc. flava. The two clusters each contain genes for AmphN homologues that form the exocyclic carboxyl group and two further cytochrome P450s for which functions cannot be predicted. The Sacc. flava structure was predicted by Usachova [31].
Usachova demonstrated production of the Sacc. flava heptaene, named flavatericin, but the partially purified material was not amenable to analysis by mass spectrometry. This heptaene had no detectable antifungal activity, as is consistent with the absence of an aminodeoxyhexose sugar. The Sacc. dendrathemae and Sacc. flava heptaenes may have other biological functions, such as signaling molecules that enable intercellular communication in mixed microbial communities [35].
The PKS genes are not completely assembled for Lentz. Waywayandensis and Lentz. Xingjiangensis. Manual curation of the fragments suggests that both clusters could encode 24-module PKSs capable of synthesizing all-trans heptaenes similar to flavatericin. The two Lentzea BGCs have AmphDIII and AmphDII homologues, indicating that the macrolactones are glycosylated with mycosamine or perosamine.
Moreover, 3-Hydroxybenzoate is predicted to serve as a primer for another class of glycosylated polyene. Holmes and co-workers identified Ps1 and Ps2 phylotypes of Pseudonocardia symbionts of ants [36]. The Ps1 phylotype strains synthesize disaccharide-modified nystatins (see below). The genomes of five Ps2 phylotype strains contain a BGC for a pentaene polyketide that is primed with a substituted benzoic acid residue. The cluster contains a Hyg5 family chorismatase (protein accession OLM23048.1) that is predicted to synthesize 3-hydroxybenzoate. The PKS starts with a CoA ligase that is predicted to activate a substituted benzoic acid. The structure predicted for the product of this BGC is shown in Figure 9. So far, it has not been possible to detect production of this pentaene [36].
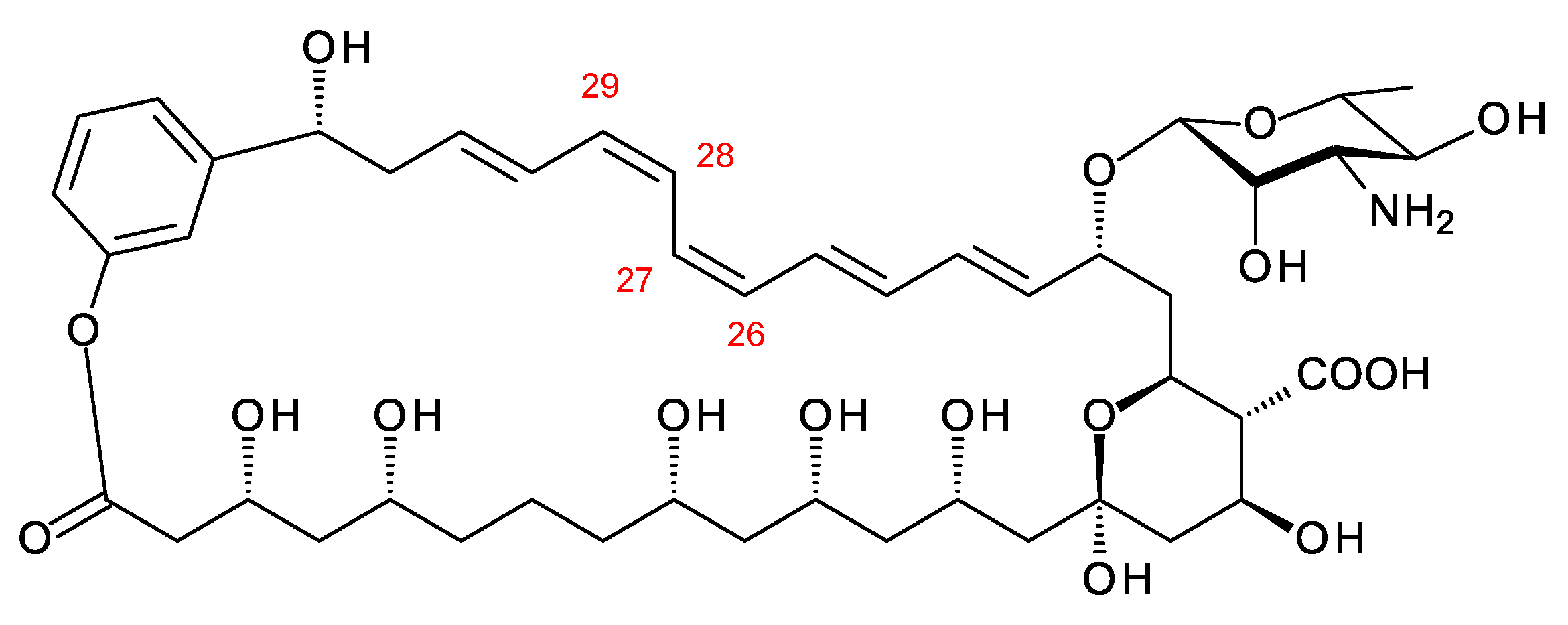
Figure 9. Ps2 phylotype polyene. This structure is the same as that predicted by Holmes and co-workers [37], except for the geometry of the C26–C27 and C28–C29 double bonds. In the PKS extension modules 3 and 4, DH domains are paired with A-type KR domains, indicating that cis alkenes are formed. The genome used for this structure prediction has the accession number MCIQ00000000.
3. Meijiemycin and Related Linear Polyene Polyols
A new class of glycosylated polyene is synthesized by Streptomyces SD50, an organism found in a marine sediment sample taken from a mangrove swamp in Singapore [38]. Meijiemycin is a linear polyene polyol (Figure 10) that is glycosylated with d-perosamine (4-amino-4,6-dideoxy-d-mannose). The biosynthetic genes are weakly expressed in wild-type Streptomyces SD50. Low and co-workers [38] increased production by inactivating two competing clusters and by growing the double mutant in the presence of mannitol, thought to boost formation of GDP-α-d-mannose, a perosamine precursor. The mode of action of meijiemycin was investigated by treating Candida albicans cells with an Alexafluor 488-labeled derivative. Examination by fluorescence microscopy revealed that the polyene localized in ergosterol-rich membrane microdomains and impaired formation of hyphal filaments from yeast cells [38]. Meijiemycin has a minimum inhibitory concentration (MIC) of 12 ± 4 μg/mL, a lower antifungal activity than cyclized polyene macrolides [38]. However, preventing the yeast to hyphal switch may abolish virulence without killing the fungal cell [39].
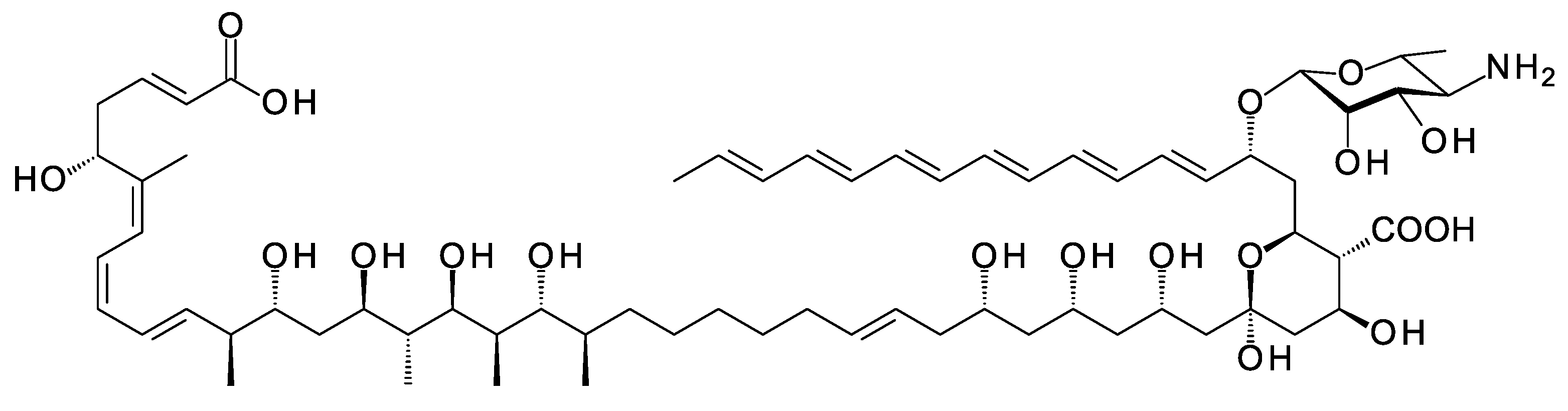
Figure 10. Structure of meijiemycin.
Amycolatopsis lexingtonensis has a BGC predicted to specify a hexaene polyol related to meijiemycin (Figure 11). AntiSMASH predicts that the polyketide chain contains methyl branches at C2, C26, and C44, and a methoxy branch at C12. AT24 introduces the C12 methoxy branch and has FAAH in place of the YASH or HAFH motifs characteristic of methylmalonate and malonate-specific AT domains. AT8 and AT29 also have FAAH motifs, raising the possibility that one or both of these domains use methoxymalonyl extenders to form methoxy branches at C44 and C2. The cluster does encode an AmphN homologue (64% identity), which would oxidize a methyl branch at C44. The genome of Amycolatopsis eburnea contains an incompletely sequenced cluster that is closely related to this hexaene-polyol BGC from Amyc. lexingtonensis.
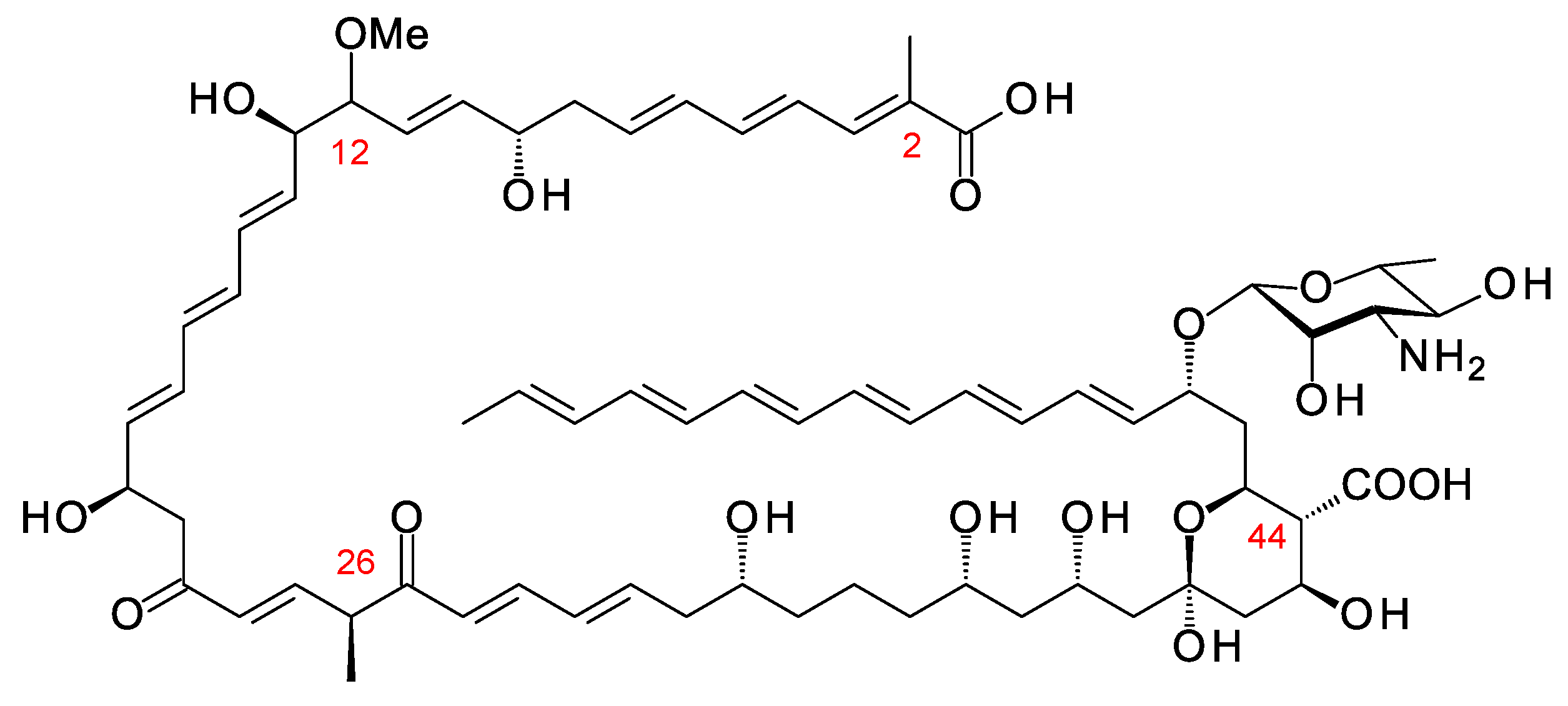
Figure 11. Predicted partial structure of Amyc. lexingtonensis polyene. Methyl branches are shown at C2 and C44 as predicted by antiSMASH, although the relevant domains AT29 and AT8 have FAAH motifs, such as the methoxymalonate-specific AT24, not YASH motifs, such as methylmalonate-specific ATs. AT17 has YASH, and so module 17 is predicted to introduce the methyl branch at C26.
Amyc. lexingtonensis has a second set of AmphDIII-DI-DII homologues associated with a BGC for a non-polyene polyketide related to vancosresmycin, which is also glycosylated with d-mycosamine [15].
Streptomyces milbemycinicus, Streptomyces bingchengensis, and Streptomyces 14.10 all have incomplete BGCs closely related to that for meijiemycin, as does a metagenomic DNA sequence from Streptomyces isolate Bin7.12.2. All of these clusters include genes for proteins that are >96% identical to the MjmSII GDP-perosamine synthase, indicating that the polyenes are glycosylated with d-perosamine.
Incomplete BGCs for meijiemycin-related polyene polyols are also present in genomes of other actinomycetes, namely Streptosporangium album DSM 43023, Acrocarpospora macrocephala, and Acrocarpospora pleiomorpha. Manual curation of the fragmented sequences indicates that these clusters specify structures that are slightly different from meijiemycin. The clusters from the two Acrocarpospora species have genes for dTDP-glucose 4,6-DH and an additional GT that is not homologous to any GT of known function.
4. Kineosporicin/Actinospene
Actinokineospora spheciospongiae DSM45935 synthesizes a methyltetraene that is glycosylated with d-perosamine, contains two epoxide groups, and is also unusual in that it is hydroxylated at C10 (Figure 12). This tetraene has been characterized independently by two groups and named kineosporicin [12] and actinospene [40]. The Actino. spheciospongiae BGC is closely related to a complete cluster in Actinokineospora mzabensis. Actino. spheciospongiae was isolated from a sponge in the Red Sea, Egypt [41], whereas Actino. mzabensis was obtained from a sample of Saharan soil taken from South Algeria [42].
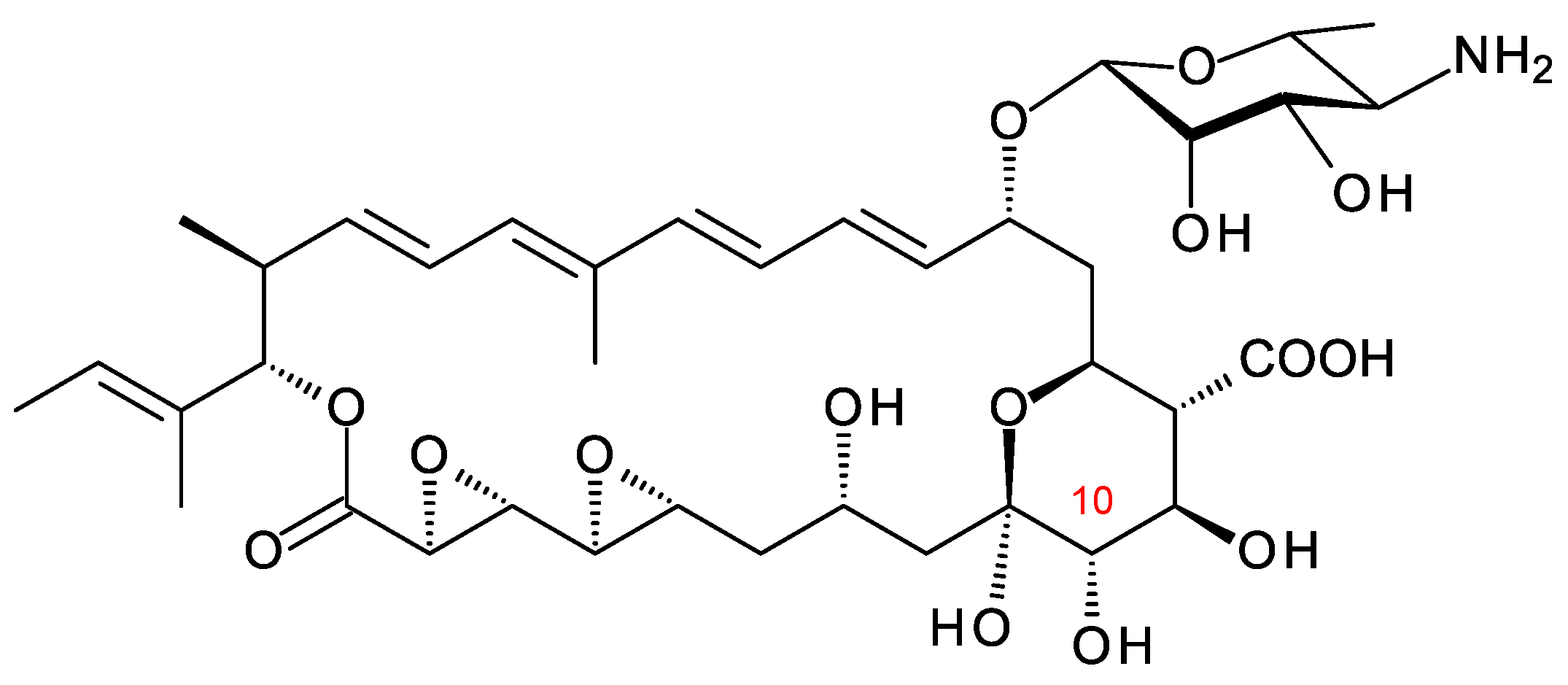
Figure 12. Kineosporicin/actinospene from A. spheciospongiae.
The Actino. spheciospongiae cluster contains genes for four cytochrome P450 enzymes. One is an AmphN homologue that forms the exocyclic carboxyl group. The other three are likely to form the two epoxides and hydroxylate C10. It is not yet known which P450 catalyzes each of these reactions. Actinospene was active against yeasts and filamentous fungi and had MICs comparable to those of pimaricin (2 to 10 μg/mL). The diepoxide and the extra C10 hydroxylation apparently do not increase antifungal activity but could have other positive effects on other pharmacological properties [12][40].
5. Enzymes Involved in Synthesis and Attachment of Mycosamine or Perosamine
Most glycosylated polyene macrolides are modified with a d-mycosamine sugar. A few are modified with d-perosamine. Perosamine also occurs in the lipopolysaccharides of some Gram-negative pathogens. The biosynthesis of GDP-α-d-perosamine from GDP-α-d-mannose has been reconstituted in vitro and occurs in two steps: GDP-α-d-mannose 4,6-dehydratase catalyzes the formation of GDP-4-keto-6-deoxy-α-d-mannose, and perosamine synthase catalyzes transamination [43] (Figure 13). It has been proposed that GDP-α-d-mycosamine formation involves isomerization of GDP-4-keto-6-deoxy-α-d-mannose to GDP-3-keto-6-deoxy-α-d-mannose, followed by a transamination catalyzed by mycosamine synthase (Figure 13). There are several 3,4-ketoisomerases that act on dTDP-4-keto-6-deoxy-α-d-glucose, but no counterpart has been identified that acts on GDP-4-keto-6-deoxy-α-d-mannose. Spontaneous 3,4-ketoisomerization of GDP-4-keto-6-deoxy-α-d-mannose occurs slowly in vitro [44], suggesting that this step in mycosamine biosynthesis proceeds in the absence of an enzyme in vivo. GDP-α-d-mycosamine is a precursor for biosynthesis of vancoresmycin, a non-polyene macrolide. The vancosresmycin biosynthetic gene cluster contains AmphDIII and AmphDII homologues but also lacks a candidate 3,4-ketoisomerase gene [15]. Fluviricin macrolactam antibiotics contain l-mycosamine, the mirror image stereoisomer of d-mycosamine [45]. Biosynthesis of dTDP-α-l-mycosamine from dTDP-4-keto-6-deoxy-α-d-glucose also requires a 3,4-ketoisomerization step for which no gene or enzyme has been identified. These observations amount to circumstantial evidence that the 3,4-ketoisomerization is not enzyme-catalyzed. However, so far, it has not been possible to synthesize GDP-α-d-mycosamine from GDP-α-d-mannose in vitro, using GDP-α-d-mannose 4,6-dehydratase and GDP-α-d-mycosamine synthase enzymes [46].
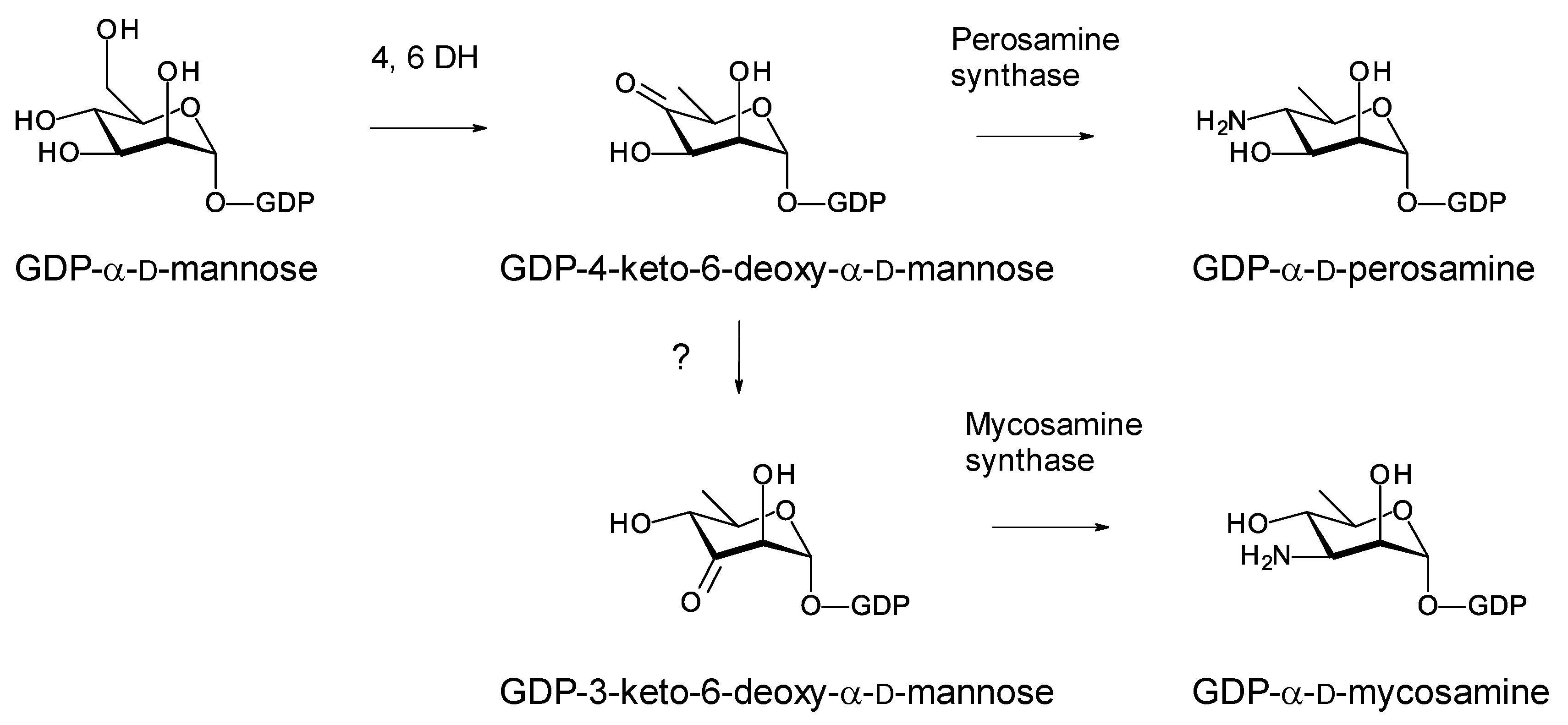
Figure 13. Biosynthetic pathways for GDP-α-d-perosamine and GDP-α-d-mycosamine.
Four classes of polyene macrolides are now known that are glycosylated with perosamine. These are perimycin, meijiemycin, kineosporicin/actinospene, and a pimaricin analogue named JB1R-13 [47]. The sequence of the BGC for JB1R-13 is not available. The BGCs for perimycin, meijiemycin, and kineosporicin provide three classes of verified GDP-α-d-perosamine synthases that function in GPM biosynthesis: (1) PerDII (Accession ADD39188.1), (2) MjmSII (Accession AYW76484.1), and (3) Actinokineospora spheciospongiae perosamine synthase (Accession WP_035287523.1). Databases contain two transaminases that are >99.72% identical to PerDII and four that are >96% identical to MjmSII.
At the primary-sequence level, these three classes of polyene GDP-α-d-perosamine synthases show 69 to 71% sequence identity to each other and are 69 to 73% identical to AmphDII, a GDP-α-d-mycosamine synthase. The alignment of these three polyene GDP-perosamine synthases with known GDP-mycosamine synthases revealed no obvious motifs or conserved residues specific for either group (unpublished results). The polyene ketosugar aminotransferases show only low homology (36 to 40% sequence identity) to perosamine synthases that function in lipopolysaccharide biosynthesis in Gram-negative bacteria [43].
Perosamine- and mycosamine-specific glycosyltransferase sequences show 57 to 62% identity, with no characteristic differences in the C-terminal GDP-sugar binding domain [48]. Primary sequence homology is not useful for bioinformatic prediction of whether a BGC specifies a polyene glycosylated with mycosamine or perosamine. The Mitchell group has devised a pHMM model for predicting whether a polyene aminosugar is perosamine or mycosamine [12]. In the case of amphotericin B, replacing the mycosamine with perosamine made little difference to antifungal activity in vitro [48].
6. Disaccharide-Modified Polyenes
Medicinal chemistry has shown that the addition of a second sugar to the mycosamine residue contributes to improvements in the pharmacological properties of amphotericin B and other polyene macrolides [49]. In dimethylformamide, the aldehyde group of glucose efficiently condenses with the amino group of mycosamine to form an imine that rearranges to an N-fructosyl analogue. A few naturally occurring polyenes have a second sugar residue linked to C4′ of mycosamine [17].
6.1. A Disaccharide-Modified Aromatic Heptaene, 67-121C
The disaccharide-modified aromatic heptaene 67-121C (Figure 14) is produced by Couchioplanes caeruleus DSM43634, formerly known as Actinoplanes caeruleus. A predicted stereostructure for the 67-121 polyketide [23] showed agreement with the experimentally determined stereostructures for members of the partricin group, except for the C41 hydroxy group (see above).
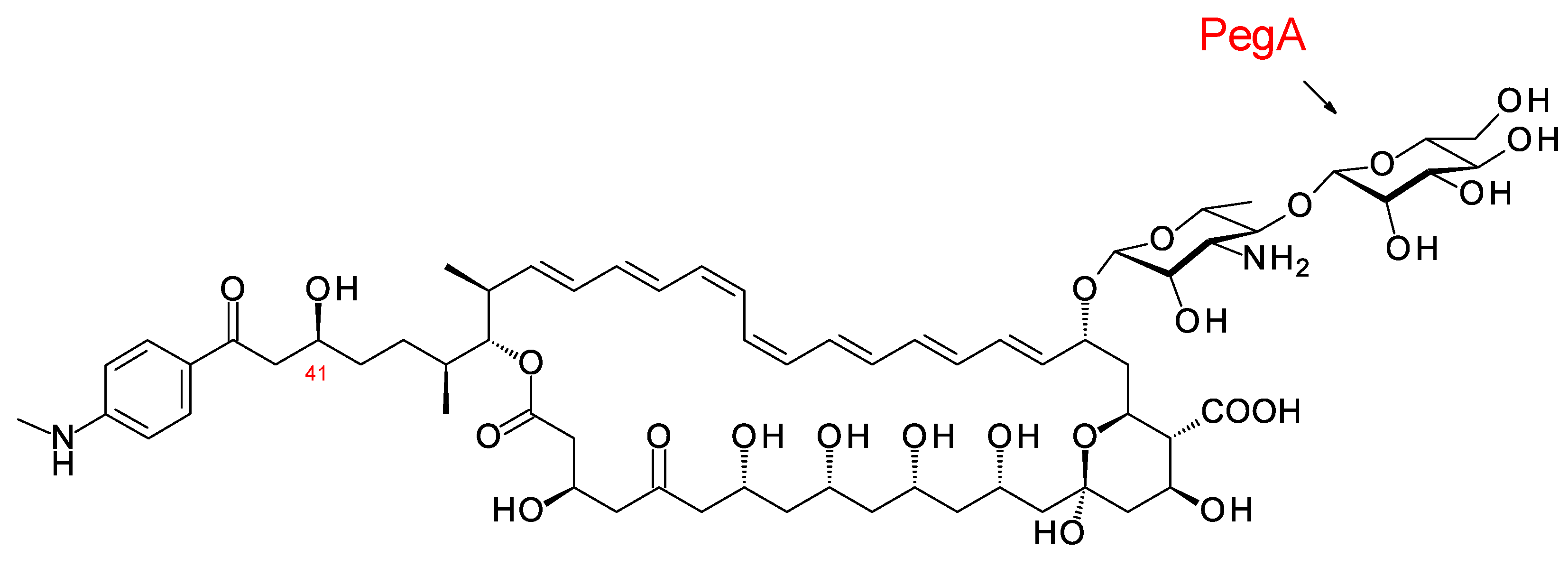
Figure 14. Structure of 67-121C.
The polyene extending glycosyltransferase PegA adds the second mannosyl sugar to the mycosamine residue of the aromatic heptaene 67-121A to give the disaccharide-modified form, 67-121C [50] (Figure 14). The function of the enzyme was confirmed by heterologous expression of the pegA gene in the candicidin producer Streptomyces albidoflavus, which gave low but detectable yields of mannosyl-candicidins [23][51]. The pegA gene is not present in the 67-121 BGC but is located at a distance of 23,437 bp along the chromosome within a transposable element. The genome of Actinoplanes digitatis has a cluster identical to the 67-121 BGC but lacks an extending glycosyltransferase (EGT) gene.
6.2. Disaccharide-Modified Nystatins
Groups in the UK and Korea discovered Pseudonocardia species that synthesize two different disaccharide-modified analogues of nystatin (Figure 15) [52][53]. In both analogues, the second sugar is attached to C4′ of mycosamine. Pseudonocardia P1 is an ant symbiont that synthesizes a nystatin in which the second sugar is mannose, added by the NypY extending GT [52]. Pseudonocardia autotrophica KCTC9441 synthesizes NPP A1 (nystatin-like Pseudonocardia polyene), in which the second sugar is N-acetylglucosamine (GlcNAc). Compared to nystatin A1, NPP A1 has a 2-fold decrease in antifungal activity but is 10 times less haemolytic and 300 times more water-soluble [53]. The extending GT is NppY. During NPP A1 biosynthesis, the addition of the second sugar precedes the final late modification, hydroxylation of the macrolactone at C10 [54]. The NppL P450 catalyzes this reaction with the disaccharide-modified 10-deoxynystatin as substrate. In Streptomyces noursei, the original nystatin A1 producer, the corresponding NysL P450 acts on a monosaccharide-modified 10-deoxytetraene substrate [55].
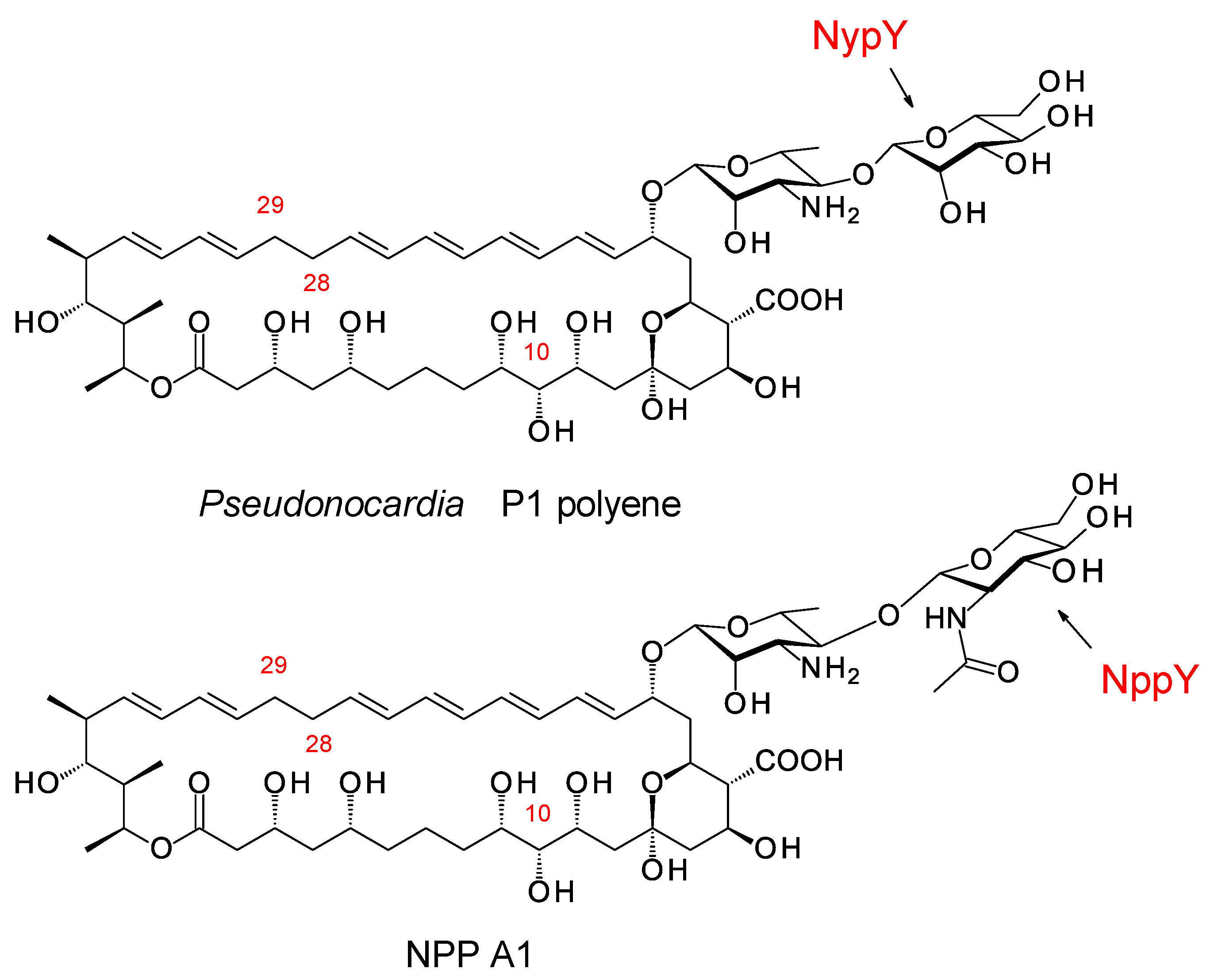
Figure 15. Disaccharide-modified Pseudonocardia polyenes.
The NppL and NysL P450s have been expressed in ΔnppL and ΔnppL-nppY mutants of Ps. autotrophica [56]. The NppL C10 hydroxylase was apparently specific for disaccharide-modified 10-deoxy macrolactones, whereas S. noursei NysL hydroxylated both monosaccharide- and disaccharide-modified 10-deoxy substrates. The in vivo complementation approach was used to assess a series of NppL–NysL hybrid enzymes. This work showed that the C-terminal 50 amino acid residue region of NppL functions in rejection of monosaccharide-modified 10-deoxy substrates [56].
Ps. autotrophica KCTC9441 has been genetically engineered extensively. This work has delivered four disaccharide-containing analogues of NPP A1: (1) a 10-deoxy tetraene, NPP A2 [57]; (2) a heptaene with a double bond between C28 and C29, NPP B1 [58]; (3) a 10-deoxy heptaene, NPP B2 [59]; and (4) NPP A3, a tetraene in which mannose replaces the GlcNAc sugar [60]. The 10-deoxy tetraene and the heptaene NPP B1 had increased antifungal activity. The 10-deoxyheptaene NPP B2 had good activity and significantly reduced hemolytic activity. Replacing the GlcNAc of NPP A1 with mannose was detrimental, as the NPP A3 analogue was inferior in terms of antifungal activity and hepatotoxicity [60]. This indicates that the structure of the second sugar is important.
Yields of the potentially improved NPP B1 and NPP B2 analogues were increased by strain improvement of the producer organisms. This was achieved by nitrosoguanidine mutagenesis and screening for increased production, and by eliminating a 128 kb plasmid carrying a BGC for a competing polyketide. In addition, an integrating plasmid was used to introduce an extra copy of the 32 kb region containing positive regulatory genes. These measures increased the yields to over 31 mg per liter of production culture for NPP B1 [61] and 7 mg per liter for NPP B2 [59].
6.3. Identification of New Polyene Extending GTs by Genome Mining
Homologues of NypY and NppY occur in genomes of several Pseudonocardia species. A protein identical to NppY is encoded within the genome of Ps. autotrophica DSM43083, which was sequenced because of its lignolytic activity [62]. NppY is also encoded within the genome of Pseudonocardia SID8383, which was sequenced as part of a project aimed at characterizing antibiotic-producing symbionts of ants [63]. Two slightly different proteins that are both 99% identical to NppY are encoded within the genomes of an endophyte Pseudonocardia alni [64] and an Antarctic isolate Pseudonocardia antarctica [65]. Another ant symbiont, Ps AL041005-10, has an enzyme that is 89% identical to NppY [66].
Proteins that are >99% identical to the mannose-specific NypY occur in the genomes of several ant symbionts. An identical protein is encoded within Ps. Ae707Ps1 [36]. Genes for a protein with 99% sequence identity occur within the genomes of Pseudonocardia strains ECO80610-09 and ECO80619-01 [67]. A slightly different protein, also 99% identical to NypY, is encoded within the genomes of four other strains, namely Ae150Aps1, Ae168Ps1, Ae263Ps1, and Ae356Ps1 [36].
In summary, six Pseudonocardia genomes have a gene for a protein that is at least 89% identical to NppY, and eight genomes have a gene for a protein that is at least 99% identical to NypY . All of these GTs are encoded by genes in nystatin BGCs. Alignment of NppY with NypY reveals 83% sequence identity. Alignment of the variants of these enzymes reveal few differences between N-acetylglucosamine-specific and mannose-specific enzymes. Extending GTs share about 50% sequence identity with their corresponding mycosaminyltransferases, but sequence alignments reveal clear sequence differences between the two groups. Some amino acid residues and motifs characteristic of NppY and NypY are also conserved in PegA. During manual curation of a polyene BGC, it is possible to distinguish between an enzyme that mycosaminylates a polyene aglycone and an extending GT that adds a second sugar to the mycosamine. It was found that two new GT sequences that align with PegA. These are encoded within polyene BGCs in Cryptosporangium arvum DSM 44712 and Amycolatopsis suaedae.
Crypto. arvum DSM 44712 is a Japanese soil isolate [68] containing a BGC for an octaene (Figure 16). Two of the PKS genes have frameshifts that may be sequencing errors or real mutations. The cluster includes a gene for a putative extending GT (EXG82143.1) that is homologous (49% identical and 64% similar) to PegA (WP_071803650.1). This polyene is of interest because the Zotchev group found that a minor octaene analogue of nystatin had high antifungal activity [69].
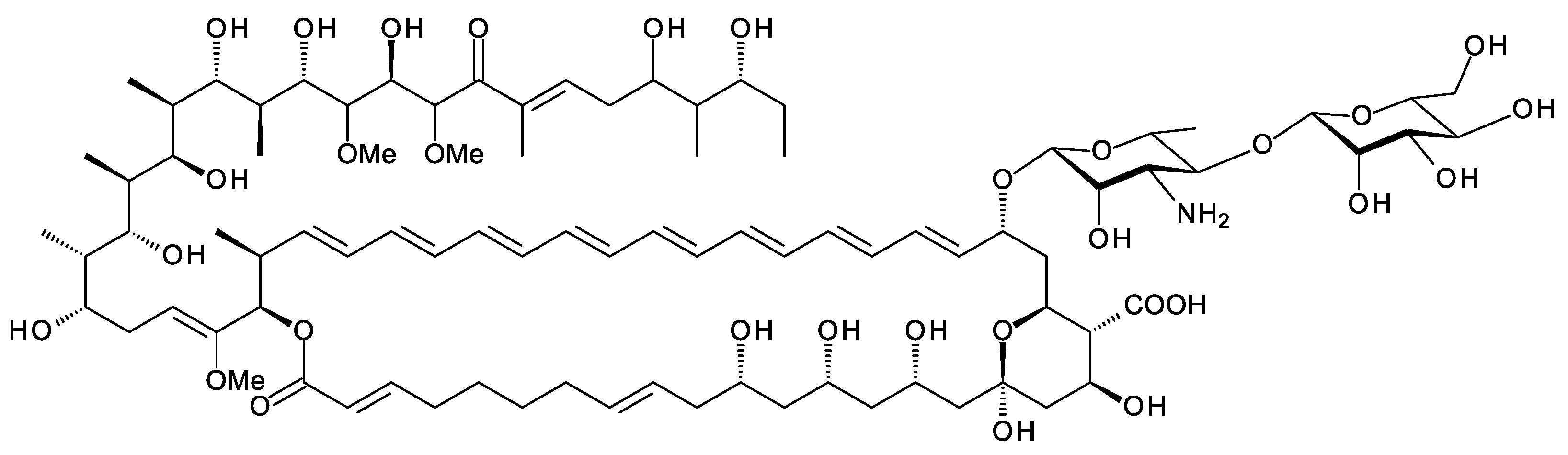
Figure 16. Predicted structure for octaene from Cryptosporangium arvum.
Amyc. suaedae is an endophyte obtained from a Suaeda maritima salt marsh plant collected in Thailand [70]. The Amyc. suaedae genome has a BGC for a methyl tetraene with a long side-chain (Figure 17). An AmphDI homologue (WP_1304475493.1, 55% identity) is likely to add the mycosamine residue. There is a gene for an extending glycosyltransferase (WP_130478880.1) that is 59% identical to PegA. This is predicted to mannosylate the mycosamine at C4′. The cluster contains genes for biosynthesis and attachment of additional deoxysugars (see below), suggesting that the real polyene may be more extensively glycosylated than the hypothetical core structure in Figure 17.
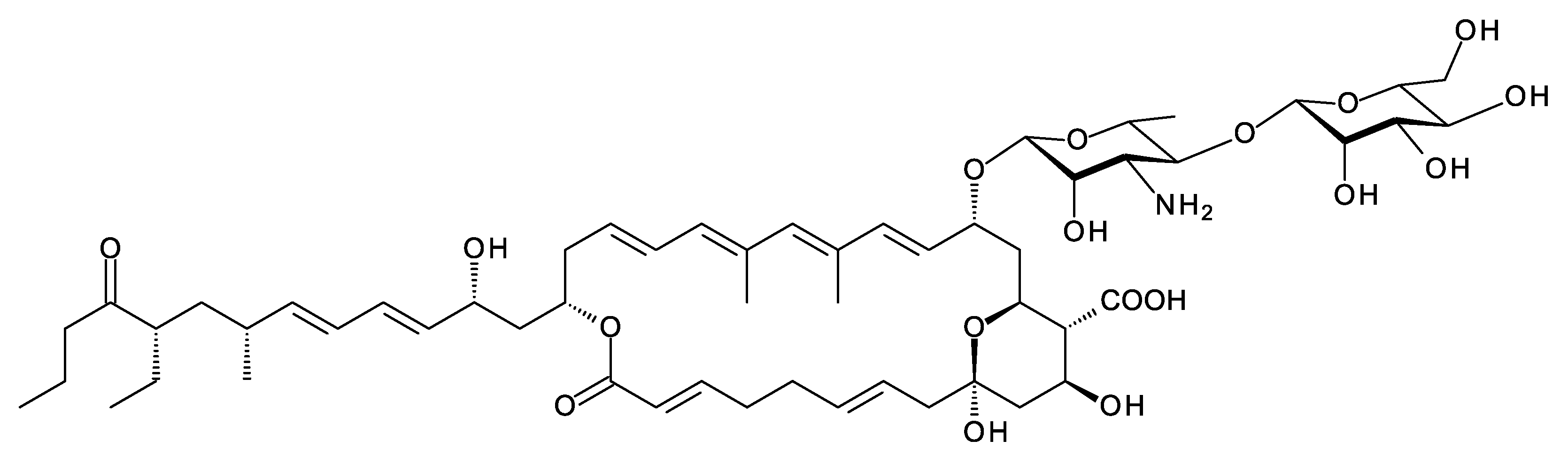
Figure 17. Partial structure predicted for Amyc. suaedae tetraene. The BGC contains genes for a AmphN P450 homologue (63% identity) that forms the exocyclic carboxyl group and genes for two further cytochrome P450 enzymes (WP_130475496.1 and WP_130475500.1) homologous to SelL (47% identity) and Lcm10 (47% identity). The modifications catalyzed by these enzymes cannot be predicted. Lcm10 catalyzes epoxide formation in lucensomycin. There are also genes for biosynthesis of additional deoxyhexoses and two further GT genes.
6.4. Extending GTs in Synthetic Biology
Early studies have investigated the potential of extending glycosyltransferases (EGTs) as tools in synthetic biology. NypY has been expressed in the amphotericin producer, Streptomyces nodosus, and in engineered strains that synthesize various analogues [51]. NypY gave modest mannosylation of amphotericins A and B and 7-ketoamphotericins A and B, but not the less toxic analogues in which a methyl group replaces the exocyclic carboxyl group. Expression in S. nodosus ΔamphL gave mannosyl-8-deoxyamphotericins A and B [71]. The yields of these analogues were higher than for C8-hydroxylated disaccharide-modified forms. This suggests that NypY is specific for 10-deoxynystatins and 8-deoxyamphotericins but does not efficiently glycosylate forms that are hydroxylated at C10 or C-8. Expression of NypY in S. noursei did not give disaccharide-modified NPP A1 [72], possibly because the native pathway rapidly C-10 hydroxylates and exports the 10-deoxynystatin A1 substrate before NypY has time to act.
Mannosyl-8-deoxyamphotericin B has been purified and characterized by NMR spectroscopy. The second sugar gave no improvement in in vitro antifungal activity compared to 8-deoxyamphotericin B, and a slight reduction in hemolytic activity [71].
NypY shows a greater tolerance towards unnatural acceptor substrates than PegA. NypY acts on candicidins, as well as some amphotericins, whereas PegA acts on candicidins but not amphotericins [51]. These studies indicate that extending GTs have potential for synthesizing disaccharide analogues of amphotericins, candicidins, and possibly other polyenes, such as pimaricin.
References
- Weissman, K.J. Polyketide stereocontrol: A study in chemical biology. Beilstein J. Org. Chem. 2017, 13, 348–371.
- Del Vecchio, F.; Petkovic, H.; Kendrew, S.G.; Low, L.; Wilkinson, B.; Lill, R.; Cortes, J.; Rudd, B.A.M.; Staunton, J.; Leadlay, P.F. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 2003, 30, 489–494.
- Caffrey, P. Conserved Amino Acid Residues Correlating with Ketoreductase Stereospecificity in Modular Polyketide Synthases. ChemBioChem 2003, 4, 654–657.
- Keatinge-Clay, A.T. A Tylosin Ketoreductase Reveals How Chirality Is Determined in Polyketides. Chem. Biol. 2007, 14, 898–908.
- Keatinge-Clay, A.T. Stereocontrol within polyketide assembly lines. Nat. Prod. Rep. 2016, 33, 141–149.
- Kwan, D.H.; Sun, Y.; Schulz, F.; Hong, H.; Popovic, B.; Sim-Stark, J.C.; Haydock, S.F.; Leadlay, P.F. Prediction and Manipulation of the Stereochemistry of Enoylreduction in Modular Polyketide Synthases. Chem. Biol. 2008, 15, 1231–1240.
- Caffrey, P.; Aparicio, J.; Malpartida, F.; Zotchev, S. Biosynthetic Engineering of Polyene Macrolides Towards Generation of Improved Antifungal and Antiparasitic Agents. Curr. Top. Med. Chem. 2008, 8, 639–653.
- Kadota, I.; Hu, Y.; Packard, G.K.; Rychnovsky, S.D. A unified approach to polyene macrolides: Synthesis of candidin and nystatin polyols. Proc. Natl. Acad. Sci. USA 2004, 101, 11992–11995.
- Aparicio, J.; Caffrey, P.; Gil, J.A.; Zotchev, S. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2002, 61, 179–188.
- Caffrey, P.; Lynch, S.; Flood, E.; Finnan, S.; Oliynyk, M. Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem. Biol. 2001, 8, 713–723.
- Liang, D.; Yang, X.; Liu, J.; Caiyin, Q.; Zhao, G.; Li, L.; Qiao, J. Global evolution of glycosylated polyene macrolide antibiotic biosynthesis. Mol. Phylogenetics Evol. 2018, 127, 239–247.
- Guo, X.; Zhang, J.; Li, X.; Xiao, E.; Lange, J.D.; Rienstra, C.M.; Burke, M.D.; Mitchell, D.A. Sterol Sponge Mechanism Is Conserved for Glycosylated Polyene Macrolides. ACS Cent. Sci. 2021, 7, 781–791.
- Amaya, J.A.; Lamb, D.C.; Kelly, S.L.; Caffrey, P.; Murarka, V.C.; Poulos, T.L. Structural Analysis of P450 AmphL from S. nodosus Provides Insights into Substrate Selectivity of Polyene Macrolide Antibiotic Biosynthetic P450s. J. Biol. Chem. 2022.
- Kells, P.M.; Ouellet, H.; Santos-Aberturas, J.; Aparicio, J.F.; Podust, L.M. Structure of Cytochrome P450 PimD Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs via a Hydroperoxoferric Intermediate. Chem. Biol. 2010, 17, 841–851.
- Kepplinger, B.; Morton-Laing, S.; Seistrup, K.H.; Marrs, E.C.L.; Hopkins, A.; Perry, J.D.; Strahl, H.; Hall, M.; Errington, J.; Allenby, N.E.E. Mode of Action and Heterologous Expression of the Natural Product Antibiotic Vancoresmycin. ACS Chem. Biol. 2018, 13, 207–214.
- Spindler, S.; Wingen, L.M.; Schönenbroicher, M.; Seul, M.; Adamek, M.; Essig, S.; Kurz, M.; Ziemert, N.; Menche, D. Modular Fragment Synthesis and Bioinformatic Analysis Propose a Revised Vancoresmycin Stereoconfiguration. Org. Lett. 2021, 23, 1175–1180.
- Caffrey, P.; De Poire, E.; Sheehan, J.; Sweeney, P. Polyene macrolide biosynthesis in streptomycetes and related bacteria: Recent advances from genome sequencing and experimental studies. Appl. Microbiol. Biotechnol. 2016, 100, 3893–3908.
- Yushchuk, O.; Ostash, I.; Mösker, E.; Vlasiuk, I.; Deneka, M.; Rückert, C.; Busche, T.; Fedorenko, V.; Kalinowski, J.; Süssmuth, R.D.; et al. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 1–14.
- McAlpine, J.B.; Bachmann, B.O.; Piraee, M.; Tremblay, S.; Alarco, A.-M.; Zazopoulos, A.E.; Farnet, C.M. Microbial Genomics as a Guide to Drug Discovery and Structural Elucidation: ECO-02301, a Novel Antifungal Agent, as an Example. J. Nat. Prod. 2005, 68, 493–496.
- Hong, H.; Fill, T.; Leadlay, P.F. A Common Origin for Guanidinobutanoate Starter Units in Antifungal Natural Products. Angew. Chem. Int. Ed. 2013, 52, 13096–13099.
- Hong, H.; Samborskyy, M.; Usachova, K.; Schnatz, K.; Leadlay, P.F. Sulfation and amidinohydrolysis in the biosynthesis of giant linear polyenes. Beilstein J. Org. Chem. 2017, 13, 2408–2415.
- Zhai, G.; Wang, W.; Xu, W.; Sun, G.; Hu, C.; Wu, X.; Cong, Z.; Deng, L.; Shi, Y.; Leadlay, P.F.; et al. Cross-Module Enoylreduction in the Azalomycin F Polyketide Synthase. Angew. Chem. Int. Ed. 2020, 59, 22738–22742.
- Sheehan, J.; Murphy, C.D.; Caffrey, P. New insights into polyene macrolide biosynthesis in Couchioplanes caeruleus. Mol. BioSyst. 2017, 13, 866–873.
- Hou, J. Mechanisms Underlying Polyene Macrolide Mediated Rescue of Growth in Ion Channel Deficient Yeast. Ph.D. Thesis, University of Illinois at Urbana-Champaign, Urbana, IL, USA, 2018. Available online: https://www.ideals.illinois.edu/handle/2142/101766 (accessed on 28 January 2022).
- Vieira, D.B. Cationic lipids and surfactants as antifungal agents: Mode of action. J. Antimicrob. Chemother. 2006, 58, 760–767.
- Floss, H.G.; Yu, T.-W.; Arakawa, K. The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: A review. J. Antibiot. 2010, 64, 35–44.
- Kang, Q.; Shen, Y.; Bai, L. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012, 29, 243–263.
- Castro, J.F.; Razmilic, V.; Gomez-Escribano, J.P.; Andrews, B.; Asenjo, J.; Bibb, M.J. Identification and Heterologous Expression of the Chaxamycin Biosynthesis Gene Cluster from Streptomyces leeuwenhoekii. Appl. Environ. Microbiol. 2015, 81, 5820–5831.
- Zhang, G.; Wang, L.; Li, J.; Zhou, Y. Amycolatopsis albispora sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 3860–3864.
- Guerrero-Garzón, J.F.; Madland, E.; Zehl, M.; Singh, M.; Rezaei, S.; Aachmann, F.L.; Courtade, G.; Urban, E.; Rückert, C.; Busche, T.; et al. Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin. iScience 2020, 23, 101785.
- Usachova, K. Analysis and Engineering of Polyene Biosynthesis in Actinomycetes. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2019.
- Andexer, J.N.; Kendrew, S.G.; Nur-E-Alam, M.; Lazos, O.; Foster, T.A.; Zimmermann, A.-S.; Warneck, T.D.; Suthar, D.; Coates, N.J.; Koehn, F.E.; et al. Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc. Natl. Acad. Sci. USA 2011, 108, 4776–4781.
- Hubrich, F.; Juneja, P.; Muller, M.; Diederichs, K.; Welte, W.; Andexer, J.N. Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold. J. Am. Chem. Soc. 2015, 137, 11032–11037.
- Zhang, C.; Moretti, R.; Jiang, J.; Thorson, J.S. The in vitro Characterization of Polyene Glycosyltransferases AmphDI and NysDI. ChemBioChem 2008, 9, 2506–2514.
- Lopez, D.; Fischbach, M.; Chu, F.; Losick, R.; Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2009, 106, 280–285.
- Holmes, N.; Innocent, T.M.; Heine, D.; Al Bassam, M.; Worsley, S.F.; Trottmann, F.; Patrick, E.H.; Yu, D.W.; Murrell, J.C.; Schiøtt, M.; et al. Genome Analysis of Two Pseudonocardia Phylotypes Associated with Acromyrmex Leafcutter Ants Reveals Their Biosynthetic Potential. Front. Microbiol. 2016, 7, 2073.
- Borzyszkowska-Bukowska, J.; Górska, J.; Szczeblewski, P.; Laskowski, T.; Gabriel, I.; Jurasz, J.; Kozłowska-Tylingo, K.; Szweda, P.; Milewski, S. Quest for the Molecular Basis of Improved Selective Toxicity of All-Trans Isomers of Aromatic Heptaene Macrolide Antifungal Antibiotics. Int. J. Mol. Sci. 2021, 22, 10108.
- Low, Z.J.; Xiong, J.; Xie, Y.; Ma, G.-L.; Saw, H.; Tran, H.T.; Wong, S.L.; Pang, L.M.; Fong, J.; Lu, P.; et al. Discovery, biosynthesis and antifungal mechanism of the polyene-polyol meijiemycin. Chem. Commun. 2020, 56, 822–825.
- Vij, R.; Hube, B.; Brunke, S. Uncharted territories in the discovery of antifungal and antivirulence natural products from bacteria. Comput. Struct. Biotechnol. J. 2021, 19, 1244–1252.
- Tang, Y.; Zhang, C.; Cui, T.; Lei, P.; Guo, Z.; Wang, H.; Liu, Q. The Discovery of Actinospene, a New Polyene Macrolide with Broad Activity against Plant Fungal Pathogens and Pathogenic Yeasts. Molecules 2021, 26, 7020.
- Kämpfer, P.; Glaeser, S.P.; Busse, H.-J.; Abdelmohsen, U.R.; Ahmed, S.; Hentschel, U. Actinokineospora spheciospongiae sp. nov., isolated from the marine sponge Spheciospongia vagabunda. Int. J. Syst. Evol. Microbiol. 2015, 65, 879–884.
- Aouiche, A.; Bouras, N.; Mokrane, S.; Zitouni, A.; Schumann, P.; Spröer, C.; Sabaou, N.; Klenk, H.-P. Actinokineospora mzabensis sp. nov., a novel actinomycete isolated from Saharan soil. Antonie Leeuwenhoek 2014, 107, 291–296.
- Albermann, C.; Piepersberg, W. Expression and identification of the RfbE protein from Vibrio cholerae O1 and its use for the enzymatic synthesis of GDP-D-perosamine. Glycobiology 2001, 11, 655–661.
- Sullivan, F.X.; Kumar, R.; Kriz, R.; Stahl, M.; Xu, G.-Y.; Rouse, J.; Chang, X.-J.; Boodhoo, A.; Potvin, B.; Cumming, D.A. Molecular Cloning of Human GDP-mannose 4,6-Dehydratase and Reconstitution of GDP-fucose Biosynthesis in Vitro. J. Biol. Chem. 1998, 273, 8193–8202.
- Miyanaga, A.; Hayakawa, Y.; Numakura, M.; Hashimoto, J.; Teruya, K.; Hirano, T.; Shin-Ya, K.; Kudo, F.; Eguchi, T. Identification of the Fluvirucin B2 (Sch 38518) Biosynthetic Gene Cluster from Actinomadura fulva subsp. indica ATCC 53714: Substrate Specificity of the β-Amino Acid Selective Adenylating Enzyme FlvN. Biosci. Biotechnol. Biochem. 2016, 80, 935–941.
- Nedal, A.; Sletta, H.; Brautaset, T.; Borgos, S.E.F.; Sekurova, O.N.; Ellingsen, T.E.; Zotchev, S.B. Analysis of the Mycosamine Biosynthesis and Attachment Genes in the Nystatin Biosynthetic Gene Cluster of Streptomyces noursei ATCC. Appl. Environ. Microbiol. 2007, 73, 7400–7407.
- Komaki, H.; Izumikawa, M.; Ueda, J.-Y.; Nakashima, T.; Khan, S.; Takagi, M.; Shin-Ya, K. Discovery of a pimaricin analog JBIR-13, from Streptomyces bicolor NBRC 12746 as predicted by sequence analysis of type I polyketide synthase gene. Appl. Microbiol. Biotechnol. 2009, 83, 127–133.
- Hutchinson, E.; Murphy, B.; Dunne, T.; Breen, C.; Rawlings, B.; Caffrey, P. Redesign of Polyene Macrolide Glycosylation: Engineered Biosynthesis of 19-(O)-Perosaminyl- Amphoteronolide B. Chem. Biol. 2010, 17, 174–182.
- Grzybowska, J.; Sowinski, P.; Gumieniak, J.; Zieniawa, T.; Borowski, E. N-Methyl-N-D-fructopyranosylamphotericin B Methyl Ester, New Amphotericin B Derivative of Low Toxicity. J. Antibiot. 1997, 50, 709–711.
- Stephens, N.; Rawlings, B.; Caffrey, P. Versatility of Enzymes Catalyzing Late Steps in Polyene 67-121C Biosynthesis. Biosci. Biotechnol. Biochem. 2013, 77, 880–883.
- De Poire, E.; Stephens, N.; Rawlings, B.; Caffrey, P. Engineered Biosynthesis of Disaccharide-Modified Polyene Macrolides. Appl. Environ. Microbiol. 2013, 79, 6156–6159.
- Barke, J.; Seipke, R.F.; Grüschow, S.; Heavens, D.; Drou, N.; Bibb, M.J.; Goss, R.J.; Yu, D.W.; Hutchings, M.I. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010, 8, 109.
- Lee, M.-J.; Kong, D.; Han, K.; Sherman, D.H.; Bai, L.; Deng, Z.; Lin, S.; Kim, E.-S. Structural analysis and biosynthetic engineering of a solubility-improved and less-hemolytic nystatin-like polyene in Pseudonocardia autotrophica. Appl. Microbiol. Biotechnol. 2012, 95, 157–168.
- Kim, H.-J.; Kim, M.-K.; Lee, M.-J.; Won, H.-J.; Choi, S.-S.; Kim, E.-S. Post-PKS Tailoring Steps of a Disaccharide-Containing Polyene NPP in Pseudonocardia autotrophica. PLoS ONE 2015, 10, e0123270.
- Volokhan, O.; Sletta, H.; Ellingsen, T.E.; Zotchev, S.B. Characterization of the P450 Monooxygenase NysL, Responsible for C-10 Hydroxylation during Biosynthesis of the Polyene Macrolide Antibiotic Nystatin in Streptomyces noursei. Appl. Environ. Microbiol. 2006, 72, 2514–2519.
- Kim, M.-K.; Won, H.-J.; Kim, H.-J.; Choi, S.-S.; Lee, H.-S.; Kim, P.; Kim, E.-S. Carboxyl-terminal domain characterization of polyene-specific P450 hydroxylase in Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 2016, 43, 1625–1630.
- Won, H.-J.; Kim, H.-J.; Jang, J.-Y.; Kang, S.-H.; Choi, S.-S.; Kim, E.-S. Improved recovery and biological activities of an engineered polyene NPP analogue in Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 2017, 44, 1293–1299.
- Kim, H.-J.; Han, C.-Y.; Park, J.-S.; Oh, S.-H.; Kang, S.-H.; Choi, S.-S.; Kim, J.-M.; Kwak, J.-H.; Kim, E.-S. Nystatin-like Pseudonocardia polyene B1, a novel disaccharide-containing antifungal heptaene antibiotic. Sci. Rep. 2018, 8, 1–8.
- Park, H.-S.; Kim, H.-J.; Han, C.-Y.; Nah, H.-J.; Choi, S.-S.; Kim, E.-S. Stimulated Biosynthesis of an C10-Deoxy Heptaene NPP B2 via Regulatory Genes Overexpression in Pseudonocardia autotrophica. Front. Microbiol. 2020, 11, 19.
- Kim, H.-J.; Kang, S.-H.; Choi, S.-S.; Kim, E.-S. Redesign of antifungal polyene glycosylation: Engineered biosynthesis of disaccharide-modified NPP. Appl. Microbiol. Biotechnol. 2017, 101, 5131–5137.
- Han, C.-Y.; Jang, J.-Y.; Kim, H.-J.; Choi, S.; Kim, E.-S. Pseudonocardia strain improvement for stimulation of the di-sugar heptaene Nystatin-like Pseudonocardia polyene B1 biosynthesis. J. Ind. Microbiol. Biotechnol. 2019, 46, 649–655.
- Grumaz, C.; Rais, D.; Kirstahler, P.; Vainshtein, Y.; Rupp, S.; Zibek, S.; Sohn, K. Draft Genome Sequence of Pseudonocardia autotrophica Strain DSM 43083, an Efficient Producer of Peroxidases for Lignin Modification. Genome Announc. 2017, 5, e01562-16.
- Chevrette, M.G.; Carlson, C.M.; Ortega-Domínguez, H.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 1–11.
- Evtushenko, L.I.; Akimov, V.N.; Dobritsa, S.V.; Taptykova, S.D. A New Species of Actinomycete, Amycolata alni. Int. J. Syst. Bacteriol. 1989, 39, 72–77.
- Prabahar, V.; Dube, S.; Reddy, G.; Shivaji, S. Pseudonocardia antarctica sp. nov. an Actinomycetes from McMurdo Dry Valleys, Antarctica. Syst. Appl. Microbiol. 2004, 27, 66–71.
- Sit, C.S.; Ruzzini, A.C.; Van Arnam, E.B.; Ramadhar, T.; Currie, C.R.; Clardy, J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2015, 112, 13150–13154.
- Van Arnam, E.B.; Ruzzini, A.C.; Sit, C.S.; Currie, C.R.; Clardy, J. A Rebeccamycin Analog Provides Plasmid-Encoded Niche Defense. J. Am. Chem. Soc. 2015, 137, 14272–14274.
- Tamura, T.; Hayakawa, M.; Hatano, K. A new genus of the order Actinomycetales, Cryptosporangium gen. nov., with descriptions of Cryptosporangium arvum sp. nov. and Cryptosporangium japonicum sp. nov. Int. J. Syst. Bacteriol. 1998, 48, 995–1005.
- Bruheim, P.; Borgos, S.E.; Tsan, P.; Sletta, H.; Ellingsen, T.E.; Lancelin, J.-M.; Zotchev, S.B.; Stocker, H.; Kruse, G.; Kreckel, P.; et al. Chemical Diversity of Polyene Macrolides Produced by Streptomyces noursei ATCC 11455 and Recombinant Strain ERD44 with Genetically Altered Polyketide Synthase NysC. Antimicrob. Agents Chemother. 2004, 48, 4148–4153.
- Chantavorakit, T.; Suksaard, P.; Matsumoto, A.; Duangmal, K. Amycolatopsis suaedae sp. nov., an endophytic actinomycete isolated from Suaeda maritima roots. Int. J. Syst. Evol. Microbiol. 2019, 69, 2591–2596.
- Walmsley, S.; De Poire, E.; Rawlings, B.; Caffrey, P. Engineered biosynthesis and characterisation of disaccharide-modified 8-deoxyamphoteronolides. Appl. Microbiol. Biotechnol. 2016, 101, 1899–1905.
- Soler, L.; Caffrey, P.; McMahon, H.E. Effects of new amphotericin analogues on the scrapie isoform of the prion protein. Biochim. Biophys. Acta (BBA) Gen. Subj. 2008, 1780, 1162–1167.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
25 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





