Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muhammad Sial | -- | 1697 | 2022-07-22 00:10:33 | | | |
| 2 | Conner Chen | + 3 word(s) | 1700 | 2022-07-25 03:38:00 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Faisal, M.; Hassan, M.; Kumar, A.; Zubair, M.; Jamal, M.; Menghwar, H.; Saad, M.; Kloczkowski, A. Role of Epigenetic Factors Responsible for Hematopoietic Niche. Encyclopedia. Available online: https://encyclopedia.pub/entry/25418 (accessed on 07 February 2026).
Faisal M, Hassan M, Kumar A, Zubair M, Jamal M, Menghwar H, et al. Role of Epigenetic Factors Responsible for Hematopoietic Niche. Encyclopedia. Available at: https://encyclopedia.pub/entry/25418. Accessed February 07, 2026.
Faisal, Muhammad, Mubashir Hassan, Aman Kumar, Muhammad Zubair, Muhammad Jamal, Harish Menghwar, Muhammad Saad, Andrzej Kloczkowski. "Role of Epigenetic Factors Responsible for Hematopoietic Niche" Encyclopedia, https://encyclopedia.pub/entry/25418 (accessed February 07, 2026).
Faisal, M., Hassan, M., Kumar, A., Zubair, M., Jamal, M., Menghwar, H., Saad, M., & Kloczkowski, A. (2022, July 21). Role of Epigenetic Factors Responsible for Hematopoietic Niche. In Encyclopedia. https://encyclopedia.pub/entry/25418
Faisal, Muhammad, et al. "Role of Epigenetic Factors Responsible for Hematopoietic Niche." Encyclopedia. Web. 21 July, 2022.
Copy Citation
Hematopoietic stem cells (HSCs) reside in a specialized microenvironment in a peculiar anatomic location which regulates the maintenance of stem cells and controls its functions. The zebrafish model is widely used to study the hematopoietic system, and has helped in the identification of various hematopoietic regulators.
hematopoietic stem cells (HSCs)
hematopoietic niche
hematopoietic microenvironment
1. Introduction
Stem cell niches comprise a specialized microenvironment that promotes stem cell maintenance and regulates its function. Hematopoietic stem cells (HSCs) niches are perivascular in the spleen and bone marrow, and certain endothelial cells and stromal cells secrete factors which promote the maintenance and regulation of HSC niches [1]. Recent progress in the field helped to identify the cellular composition of hematopoietic stem and progenitor cells (HSPC), exploring the complex molecular networks that regulate the HSPC [2]. HSCs produce a variety of hematopoietic lineage cells in a specific microenvironment in bone marrow (BM) called “niche”. Multiple cells in BM contribute to HSC niche activity, and, among these, stromal cells are closely associated with vasculature [3]. The contribution of osteoblasts in HSC maintenance is still debatable, although the role of bone-derived molecules (e.g., osteopontin) or the role of bone turnover on HSC localization and function was demonstrated [4][5]. Studies have reported that the deletion of niche factors (CXCL12) or stem cell factors (SCF) from mature osteoblasts and osteoblastic progenitor cells does not lead to a reduction of HSCs in bone marrow (BM) [6][7][8]. Chimeric antigen receptor (CAR) cells express a high amount of CXCL12 and SCF, and are mainly distributed around sinusoids, and in the form of a homogenous tangled network in BM. CAR cell depletion using CXCL12 diphtheria toin receptor (DTR) results in a reduction of HSCs in BM [9]. Conditional deletion of CXCL12 from lepR-Cre marked cells mobilizes HSCs from BM to the spleen and peripheral blood, and LepR+ stromal cells around sinusoids have been shown to regulate the mobilization of HSCs pool [6]. Nes-GFP+ cells have also been identified as niche player in BM. Stromal cells within the population of Nes-GFP+ are an important source of niche factors critical for the maintenance of HSC [3][10]. Perivascular cells also express increased levels of major niche factors associated with HSCs [10][11]. When HSPCs arrive in a perivascular niche, then a group of endothelial cells (ECs) remodel and surround a single HSPC attached to a single mesenchymal stromal cell. These mesenchymal stromal cells anchor HSPCs and orient their divisions. A compound called lycorine promotes HSPC and niche interaction during development, which expands the pool of stem cells into adulthood [12]. ECs are part of the niche components. The blockade of angiogenic activity of ECs by neutralizing vascular endothelial cadherin (VE-cadherin) and vascular endothelial growth factor receptor-2 (VEGFR-2) impairs supportive function of ECs to HSCs [13]. HSC quiescence is also regulated by ECs via surface molecule E-selectin expression [14]. Conditional deletion of CXCL12 and SCF from ECs can decrease number of HSC in BM, and suggests a role of ECs in the maintenance of HSCs by producing these niche factors [6]. However, the heterogeneity of EC populations is unresolved. ECs with an increased expression of CD31 (CD31hi) and endomucin (Emcnhi), referred to as type H endothelium, are found in end-terminal arterioles connecting to sinusoids that express Kitl encoding SCF at higher levels than sinusoidal type L ECs [15]. However, the specific contribution of EC subset still requires further analyses with selective genetic deletion of SCF. Vascular permeability difference shows a differential role of ECs between sinusoids and arterioles influencing HSC niche. Due to the reduced permeability of arterial vessels which keep HSCs in low reactive oxygen species (ROS), HSCs are manifested in a quiescent state. On the contrary, more leaky sinusoids expose HSCs to blood plasma and promote a high level of ROS in HSCs, increasing the ability of differentiation and migration [16]. The whole mechanism of hematopoietic maintenance and quiescence, and factors governing its regulation at different anatomic locations of hematopoietic niches, are shown in Figure 1.
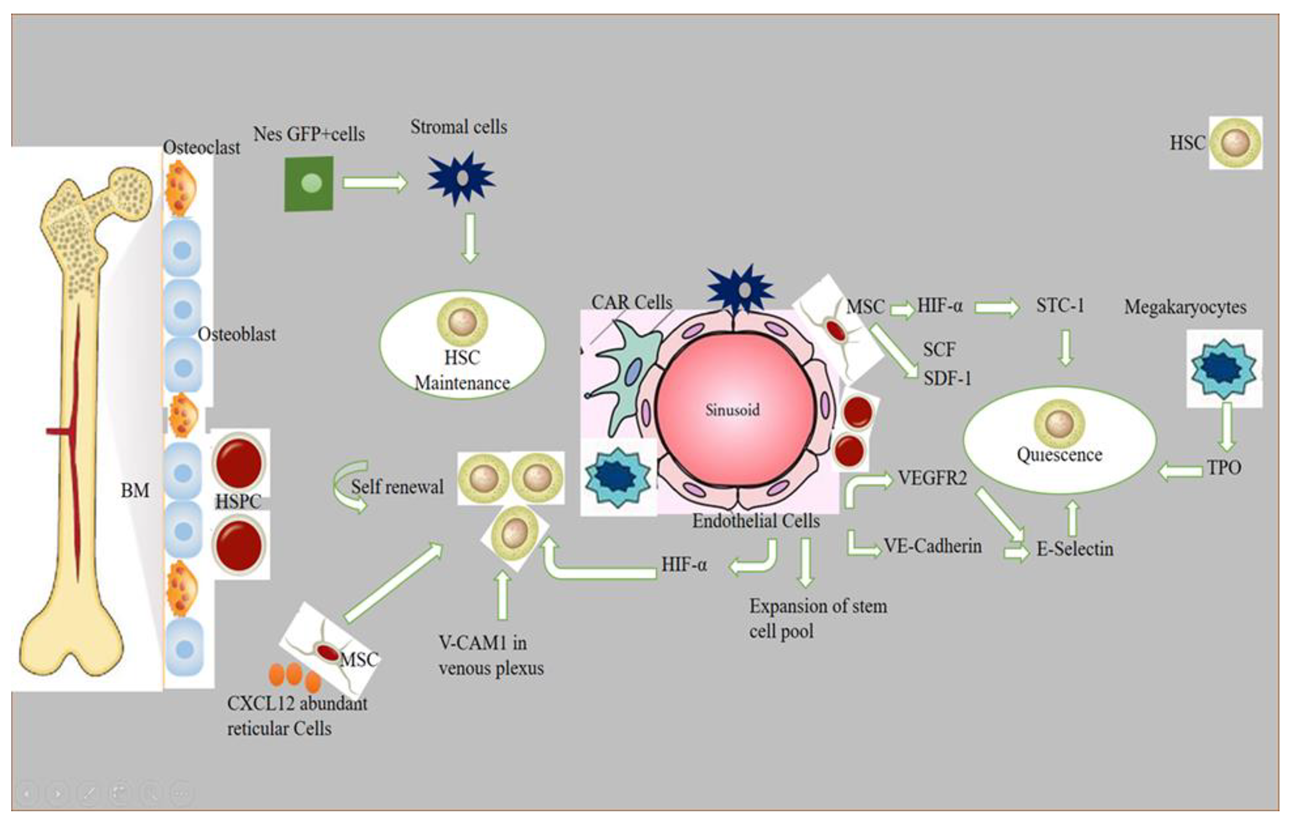
Figure 1. Role of different signaling factors responsible for hematopoietic niche maintenance and quiescence. A brief description of how normal HSPCs compete for microenvironmental space and resources. Niche cells, cytokines, signals, ECM, and oxygen gradient govern HSC activity. HSCs vary by subniche. Endosteal niches maintain LT-HSCs, while sinusoidal niches aid in hematopoietic development and regeneration. NG2+ arte-riolar pericytes block HSCs from shutting arterioles. LepR-expressing perisinusoidal cells produce SCF and CXCL12, required for HSC maintenance or mobilization. Different niches and subniches serve hematopoiesis.
Non-myelinated Schwann cells wrapping the sympathetic nerves maintain HSC quiescence by activating TGFβ. Sympathetic signals induced by granulocyte colony-forming factor (G-CSF) also play a role in HSC mobilization from niche cells [17]. Macrophages are also considered as an important element of niche-modulating cells in BM, and the deletion of macrophages have shown HSPC mobilization into blood with a reduction of niche factor encoding genes [18]. Macrophages in BM participate in the regulation of HSC through the BM microenvironment [3]. The vascular cell adhesion molecule-1 (VCAM-1) macrophage-like niche cell population in the inner surface of venous plexus interacts with HSPCs in an integrin subunit alpha 4 (ITGA4)-dependent manner, and has its role in HSPC retention within the microenvironment [19]. Megakaryocytes (MKs), if selectively depleted, lead to a loss of quiescence of HSCs, and injection of cxcl4 produced by MKs increases quiescence, which leads to HSC reduction [20]. The removal of MKs results in an increased number of HSC, and proliferation and reduction of TGF-β1 protein and nuclear-localized phosphorylated SMAD2/3 in HSCs [21]. MKs regulate HSC quiescence by producing thrombopoietin (TPO), which is a crucial cytokine for HSC quiescence, and is mediated by membrane protein C-type lectin-like receptor-2 (CLEC-2) signaling [22][23]. The use of zebrafish to study the hematopoietic niche has enabled discoveries of novel cell-to-cell interactions and important regulators of HSCs, and the mystery of niche components may contribute to therapeutic efforts to direct differentiation of HSCs to improve stem cell transplants and to sustain stem cells in culture [24].
2. Role of Epigenetic Factors Responsible for Hematopoietic Niche in Zebrafish Models
The zebrafish model is widely used to study the hematopoietic system, and has helped in the identification of various hematopoietic regulators. Recent studies on epigenetic regulation have enabled researchers to understand normal and malignant hematopoiesis [25]. Gene expression is controlled by chromatin conformation, and, if deregulated, then malignancies may occur [26]. The zebrafish serves as an excellent model to explore the mechanisms underlying chromatin regulation, and to evaluate the effects of chromatin-modifying drugs. Moreover, chromatin immunoprecipitation (chip) can be used in combination with sequencing to identify gene regulatory elements, chromatin architecture, and DNA binding sites in zebrafish [27]. Nuclear architecture protein cohesion and CCCTC binding factor (CTCF) contribute to gene regulation and chromatin structure. Cohesion is important for zygotic genome activation (ZGA). It is suggested that a subunit of cohesis Rad21, if depleted, causes a delay in ZGA; on the contrary, the depletion of CTCF affects little. Rad21 depletion destroys nucleoli formation and RNA polymerase II foci, leading to defective chromosome architecture [28]. Single cell RNA sequencing (ScRNA-seq), combined with ATAC-seq and immunophenotypic analysis, helps to integrate lineage differentiation with regulatory element accessibility [29].
Recent studies have shown that epigenetic modifications maintain hematopoietic cell fate by DNA methylation [30]. Dynamic changes in DNA methylation have been observed during cellular differentiation and development. Tissue-specific, differentially methylated regions (DMRs) overlap tissue-specific regulatory elements. The methylation pattern of developmental-stage-specific DMRs revealed a much stronger correlation than promoter methylation [31]. Hence, the developmental enhancer and DNA methylation exhibit an important status during zebrafish early development. However, the direct significance of the DNA methylation state of enhancers is unclear for most of the loci [32].
Studies have shown that epigenetic and epitranscriptomic factors are vital for reshaping gene expression patterns of hemogenic endothelial cells, as they are involved in HSC production [33]. Endothelial-to-hematopoietic transition (EHT) is required to generate HSCs, and it is brought about by transcription factors and signaling pathways, whereas Gata2 and Notch transcription factors are upstream regulators which are functionally followed by cMyb and Runx1 transcription factors [34]. These EHT genes represent DNA and RNA methylation, histone modifications, and chromatin remodeling as epigenetic mechanisms controlling the production of HSCs [33]. Moreover, epigenetic modification enables the maintenance of stem cell differentiation and development [35]. DNA methylation is an epigenetic mark, and has its role in the development of HSCs by the induction of transcriptional silencing [36][37]. Ge et al. have shown that HSC formation from the endothelium by EHT requires the ten-eleven translocation (Tet) family of cytosine dehydrogenases (tet1, tet2, Tet3), out of which, only Tet2 and Tet3 localize to the aorta gonad mesonephros (AGM) region, where HSCs bud off into the circulatory system [38]. DNA methyltransferase 1 (Dnmt1) is important in regulating gene expression by maintaining DNA methylation patterns, and it also maintains the HSPCs population in zebrafish [39].
Moreover, post-translational histone modifications in nucleosome give additional means in regulating chromatin accessibility, and it is a common mechanism to manage endothelial cells’ identity in HSC development [33][35]. Commonly, histone modifications include methylation and acetylation of lysine (K) residues on the N-terminal of the histone tail, and acetylation is carried out by histone acetyl transferases (HATs), which leads to the opening up of transcriptionally active chromatin states. On the contrary, deacetylation is carried out by histone deacetylases (HDACs), and it is responsible for gene inactivation through chromatin compaction. However, histone methylation has more effects on gene regulation that usually depend on the position of the lysine residue to be methylated [36]. Polycomb repressive complex (PRC) 1 is the earliest known epigenetic regulator of HSC formation, and works as an inhibitor for hemogenic EC specification [40]. In the later stages of HSC development, an epigenetic machinery CoREST repressive complex regulates EC identity [41].
Another type of epigenetic mechanism involved in HSC development regulation is chromatin remodeling. This process involves multi-subunit complexes which recognize the genomic landscape by ATP utilization, change the nucleosome position by sliding, and use eviction to change the nucleosome composition, followed by the reassembly of histone variants, and the resulting nucleosome compaction or expansion restricts or promotes the accessibility of transcription factors to regulatory regions [42]. Chromodomain helicase DNA-binding (CHD) is chromatin remodeling ATPase, and regulates HSC formation by the maturation of developing HSCs by increasing transcriptional output at the pro-hematopoietic gene level [33]. Overall, the role of epigenetic factors in hematopoietic niche maintenance plays a pivotal role, but further exploration is needed on the transcriptomic level and at post-transitional level to delineate the underlying mechanisms.
References
- Crane, G.; Jeffery, E.; Morrison, S. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017, 17, 573–590.
- Charbord, P.; Pouget, C.; Binder, H.; Dumont, F.; Stik, G.; Levy, P.; Allain, F.; Marchal, C.; Richter, J.; Uzan, B.; et al. A Systems Biology Approach for Defining the Molecular Framework of the Hematopoietic Stem Cell Niche. Cell Stem Cell 2014, 15, 376–391.
- Asada, N.; Takeishi, S.; Frenette, P.S. Complexity of bone marrow hematopoietic stem cell niche. Int. J. Hematol. 2017, 106, 45–54.
- Nilsson, S.K.; Johnston, H.M.; Whitty, G.A.; Williams, B.; Webb, R.J.; Denhardt, D.T.; Bertoncello, I.; Bendall, L.J.; Simmons, P.J.; Haylock, D.N. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 2005, 106, 1232–1239.
- Christodoulou, C.; Spencer, J.A.; Yeh, S.-C.A.; Turcotte, R.; Kokkaliaris, K.D.; Panero, R.; Ramos, A.; Guo, G.; Seyedhassantehrani, N.; Esipova, T.V.; et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 2020, 578, 278–283.
- Ding, L.; Morrison, S.J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 2013, 495, 231–235.
- Ding, L.; Saunders, T.; Enikolopov, G.; Morrison, S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012, 481, 457–462.
- Greenbaum, A.; Hsu, Y.-M.S.; Day, R.B.; Schuettpelz, L.G.; Christopher, M.J.; Borgerding, J.N.; Nagasawa, T.; Link, D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013, 495, 227–230.
- Omatsu, Y.; Sugiyama, T.; Kohara, H.; Kondoh, G.; Fujii, N.; Kohno, K.; Nagasawa, T. The Essential Functions of Adipo-osteogenic Progenitors as the Hematopoietic Stem and Progenitor Cell Niche. Immunity 2010, 33, 387–399.
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.; Lira, S.A.; Scadden, D.T.; Ma’Ayan, A.; Enikolopov, G.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834.
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.G.; Riminucci, M.; et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007, 131, 324–336.
- Tamplin, O.J.; Durand, E.M.; Carr, L.A.; Childs, S.; Hagedorn, E.J.; Li, P.; Yzaguirre, A.D.; Speck, N.A.; Zon, L.I. Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche. Cell 2015, 160, 241–252.
- Butler, J.M.; Nolan, D.J.; Vertes, E.L.; Varnum-Finney, B.; Kobayashi, H.; Hooper, A.T.; Seandel, M.; Shido, K.; White, I.A.; Kobayashi, M.; et al. Endothelial Cells Are Essential for the Self-Renewal and Repopulation of Notch-Dependent Hematopoietic Stem Cells. Cell Stem Cell 2010, 6, 251–264.
- Winkler, I.G.; Barbier, V.; Nowlan, B.; Jacobsen, R.N.; Forristal, C.E.; Patton, J.T.; Magnani, J.L.; Lévesque, J.-P. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 2012, 18, 1651–1657.
- Kusumbe, A.P.; Ramasamy, S.K.; Itkin, T.; Mäe, M.A.; Langen, U.H.; Betsholtz, C.; Lapidot, T.; Adams, R.H. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature 2016, 532, 380–384.
- Itkin, T.; Gur-Cohen, S.; Spencer, J.A.; Schajnovitz, A.; Ramasamy, S.K.; Kusumbe, A.P.; Ledergor, G.; Jung, Y.; Milo, I.; Poulos, M.G.; et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 2016, 532, 323–328.
- Yamazaki, S.; Ema, H.; Karlsson, G.; Yamaguchi, T.; Miyoshi, H.; Shioda, S.; Taketo, M.M.; Karlsson, S.; Iwama, A.; Nakauchi, H. Nonmyelinating Schwann Cells Maintain Hematopoietic Stem Cell Hibernation in the Bone Marrow Niche. Cell 2011, 147, 1146–1158.
- Winkler, I.G.; Sims, N.A.; Pettit, A.R.; Barbier, V.; Nowlan, B.; Helwani, F.; Poulton, I.J.; Van Rooijen, N.; Alexander, K.; Raggatt, L.J.; et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 2010, 116, 4815–4828.
- Li, D.; Xue, W.; Li, M.; Dong, M.; Wang, J.; Wang, X.; Li, X.; Chen, K.; Zhang, W.; Wu, S.; et al. VCAM-1+ macrophages guide the homing of HSPCs to a vascular niche. Nature 2018, 564, 119–124.
- Bruns, I.; Lucas, D.; Pinho, S.; Ahmed, J.; Lambert, M.P.; Kunisaki, Y.; Scheiermann, C.; Schiff, L.; Poncz, M.; Bergman, A.; et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 2014, 20, 1315–1320.
- Zhao, M.; Perry, J.M.; Marshall, H.; Venkatraman, A.; Qian, P.; He, X.C.; Ahamed, J.; Li, L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 2014, 20, 1321–1326.
- Nakamura-Ishizu, A.; Takubo, K.; Kobayashi, H.; Suzuki-Inoue, K.; Suda, T. CLEC-2 in megakaryocytes is critical for maintenance of hematopoietic stem cells in the bone marrow. J. Exp. Med. 2015, 212, 2133–2146.
- Nakamura-Ishizu, A.; Takubo, K.; Fujioka, M.; Suda, T. Megakaryocytes are essential for HSC quiescence through the production of thrombopoietin. Biochem. Biophys. Res. Commun. 2014, 454, 353–357.
- Wattrus, S.J.; Zon, L.I. Stem cell safe harbor: The hematopoietic stem cell niche in zebrafish. Blood Adv. 2018, 2, 3063–3069.
- De Pater, E.; Trompouki, E. Bloody Zebrafish: Novel Methods in Normal and Malignant Hematopoiesis. Front. Cell Dev. Biol. 2018, 6, 124.
- Gröschel, S.; Sanders, M.A.; Hoogenboezem, R.; de Wit, E.; Bouwman, B.; Erpelinck, C.; Van Der Velden, V.H.; Havermans, M.; Avellino, R.; Van Lom, K.; et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell 2014, 157, 369–381.
- Trompouki, E.; Bowman, T.V.; DiBiase, A.; Zhou, Y.; Zon, L.I. Chromatin Immunoprecipitation in Adult Zebrafish Red Cells. Methods Cell Biol. 2011, 104, 341–352.
- Meier, M.; Grant, J.; Dowdle, A.; Thomas, A.; Gerton, J.; Collas, P.; O’Sullivan, J.; Horsfield, J.A. Cohesin facilitates zygotic genome activation in zebrafish. Development 2017, 145, dev.156521.
- Buenrostro, J.D.; Corces, M.R.; Lareau, C.A.; Wu, B.; Schep, A.N.; Aryee, M.J.; Majeti, R.; Chang, H.Y.; Greenleaf, W.J. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018, 173, 1535–1548.e16.
- Gore, A.V.; Athans, B.; Iben, J.R.; Johnson, K.; Russanova, V.; Castranova, D.; Pham, V.N.; Butler, M.G.; Williams-Simons, L.; Nichols, J.T.; et al. Epigenetic regulation of hematopoiesis by DNA methylation. eLife 2016, 5, e11813.
- Lee, H.J.; Lowdon, R.F.; Maricque, B.; Zhang, B.; Stevens, M.; Li, D.; Johnson, S.L.; Wang, T. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat. Commun. 2015, 6, 6315.
- Kaaij, L.J.T.; Mokry, M.; Zhou, M.; Musheev, M.; Geeven, G.; Melquiond, A.S.J.; Domingues, A.M.D.J.; De Laat, W.; Niehrs, C.; Smith, A.D.; et al. Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol. 2016, 17, 146.
- Kasper, D.M.; Nicoli, S. Epigenetic and Epitranscriptomic Factors Make a Mark on Hematopoietic Stem Cell Development. Curr. Stem Cell Rep. 2018, 4, 22–32.
- Gritz, E.; Hirschi, K.K. Specification and function of hemogenic endothelium during embryogenesis. Experientia 2016, 73, 1547–1567.
- Atlasi, Y.; Stunnenberg, H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017, 18, 643–658.
- Li, C.; Evans, T.; Goll, M. Epigenetic regulation of hematopoietic stem cell development. Methods Cell Biol. 2016, 135, 431–448.
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017, 10, 23.
- Ge, L.; Zhang, R.-P.; Wan, F.; Guo, D.-Y.; Wang, P.; Xiang, L.-X.; Shao, J.-Z. TET2 Plays an Essential Role in Erythropoiesis by Regulating Lineage-Specific Genes via DNA Oxidative Demethylation in a Zebrafish Model. Mol. Cell. Biol. 2014, 34, 989–1002.
- Liu, X.; Jia, X.; Yuan, H.; Ma, K.; Chen, Y.; Jin, Y.; Deng, M.; Pan, W.; Chen, S.; Chen, Z.; et al. DNA methyltransferase 1 functions through C/ebpa to maintain hematopoietic stem and progenitor cells in zebrafish. J. Hematol. Oncol. 2015, 8, 15.
- Eliades, A.; Wareing, S.; Marinopoulou, E.; Fadlullah, M.Z.; Patel, R.; Grabarek, J.B.; Plusa, B.; Lacaud, G.; Kouskoff, V. The Hemogenic Competence of Endothelial Progenitors Is Restricted by Runx1 Silencing during Embryonic Development. Cell Rep. 2016, 15, 2185–2199.
- Thambyrajah, R.; Mazan, M.; Patel, R.; Moignard, V.; Stefanska, M.; Marinopoulou, E.; Li, Y.; Lancrin, C.; Clapes, T.; Möröy, T.; et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nature 2015, 18, 21–32.
- Hota, S.; Bruneau, B.G. ATP-dependent chromatin remodeling during mammalian development. Development 2016, 143, 2882–2897.
More
Information
Subjects:
Developmental Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
714
Revisions:
2 times
(View History)
Update Date:
25 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




