Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wiktoria Kasprzycka | -- | 2658 | 2022-07-19 15:44:44 | | | |
| 2 | Conner Chen | + 1 word(s) | 2659 | 2022-07-21 08:22:47 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Nowak-Stępniowska, A.; Kasprzycka, W.; Osuchowska, P.N.; Trafny, E.A.; Bartnik, A.; Fiedorowicz, H.; Wachulak, P. Soft X-ray Contact Microscopy. Encyclopedia. Available online: https://encyclopedia.pub/entry/25344 (accessed on 07 February 2026).
Nowak-Stępniowska A, Kasprzycka W, Osuchowska PN, Trafny EA, Bartnik A, Fiedorowicz H, et al. Soft X-ray Contact Microscopy. Encyclopedia. Available at: https://encyclopedia.pub/entry/25344. Accessed February 07, 2026.
Nowak-Stępniowska, Agata, Wiktoria Kasprzycka, Paulina Natalia Osuchowska, Elżbieta Anna Trafny, Andrzej Bartnik, Henryk Fiedorowicz, Przemysław Wachulak. "Soft X-ray Contact Microscopy" Encyclopedia, https://encyclopedia.pub/entry/25344 (accessed February 07, 2026).
Nowak-Stępniowska, A., Kasprzycka, W., Osuchowska, P.N., Trafny, E.A., Bartnik, A., Fiedorowicz, H., & Wachulak, P. (2022, July 20). Soft X-ray Contact Microscopy. In Encyclopedia. https://encyclopedia.pub/entry/25344
Nowak-Stępniowska, Agata, et al. "Soft X-ray Contact Microscopy." Encyclopedia. Web. 20 July, 2022.
Copy Citation
Soft X-ray microscopy is a powerful technique for imaging cells with nanometer resolution in their native state without chemical fixation, staining, or sectioning.
nanometric resolution
soft X-ray contact microscopy
cell imaging
1. Soft X-ray Contact Microscopy
The principle of soft X-ray contact microscopy is well known. It can be considered a variant of X-ray contact microradiography, in which the shadow image of a specimen is obtained by irradiating the sample on the photographic plate with X-rays[1][2]. Soft X-rays in the ”water window” spectral range, produced with a specially designed X-ray tube and fine-grained photographic emulsion, have been used for imaging cellular structures with a sub-micrometer resolution. This spatial resolution corresponds to the resolution of the light microscope system utilized in this experiment to inspect the micro radiograms[3][4]. Resolution in the nanometer range was achieved when a photoresist was used to record the microradiogram instead of a photographic plate and electron microscopy to examine the microradiogram in place of an optical microscope. This SXCM technique, based on X-ray photoresist, was adapted from X-ray lithography techniques and was quickly recognized as a promising microscopy technique for examining biological specimens[5][6]. The unique properties of photons at soft-X-ray wavelength make it possible to image cells based on the organic composition of unstained subcellular structures. X-ray techniques, including SXCM, enable the study of the remodeling of cell and extracellular matrix (ECM) architecture during normal and disease processes. The investigation of single focal adhesion (FA) in cancer, a crucial evaluation of metastatic potential, is possible thanks to SXCM.
The soft X-ray contact microscopy of biological objects with the nanometric resolution was demonstrated for the first time using an X-ray tube and synchrotron operating in the “water window” range[7][8].
The SXCM technique is schematically presented in Figure 1. It uses a three-step procedure. In the first step, called the exposure (Figure 1a), soft X-ray (SXR) radiation emitted from the source is transmitted through the sample to expose a high-resolution recording medium that is “in contact” with the sample. A thin layer of X-ray resist spun on a silicon wafer is usually used as the recording medium. It allows the recording of the local intensity modulation of radiation transmitted through the sample in its structure. The absorbed radiation dose is then converted into the change in the recording-medium morphology, creating an imprint (relief) structure in the recording medium. This conversion is achieved during the development process, a standard chemical procedure (Figure 1b). Finally, the radiation-induced changes in the recording medium can be digitized, providing a digital image (Figure 1c).
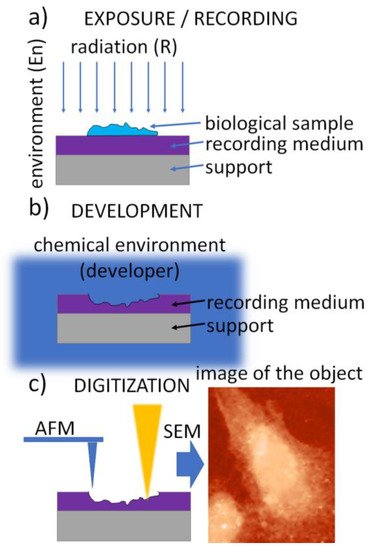
Figure 1. A flowchart of the contact X-ray microscopy imaging procedure: (a) exposure with radiation penetrating the biological sample and absorbed in the recording medium—photoresist, (b) chemical development of the photoresist converting spatial 2D modulation of the radiation density into the surface morphology, (c) digitization—conversion of the surface morphology into the digital image using high-resolution imaging methods, e.g., AFM or SEM.
To provide superior resolution compared to optical methods, the final step (Figure 1c) is performed using atomic force microscopy (AFM), scanning electron microscopy (SEM), or transmission electron microscopy (TEM); however, fluorescence microscopes (FM) are also used when lithium fluoride crystals are used as the recording medium.
2. Radiation Sources
Various types of X-ray sources in the “water window” were used in SXCM, including, X-ray tubes, synchrotrons, and plasma sources. X-ray tubes and synchrotrons were used in the early phase of SXCM research[7][9]. The main disadvantage of using these sources in SXCM was the exposure of the sample for a relatively long time, leading to radiation damage and decreased resolution[10]. It was found that imaging hydrated, living biological specimens required exposure with a sufficient X-ray dose in a very short time to avoid blurring due to radiation-induced and natural movement, as well as the recording of an image of the sample before radiation damage occurred. Therefore, laser-produced plasma sources emitting soft X-ray radiation in the form of nanosecond pulses were proposed[11] and came to dominate the use of SXCM.
A dense and high-temperature (106 K) plasma from the interaction of intense laser pulses with a target material (usually solid) is a bright source of soft X-rays. By choosing suitable target materials and laser parameters, the emitted radiation can be optimized for “water window” X-rays[12]. SXCM experiments were performed using various laser systems, including Nd:glass[13], Nd:YAG[11], iodine, excimer[14], and others, producing sub- and nanosecond pulses with energies varying from less than a Joule to several hundred Joules. The latter made it possible to obtain SXCM microscopy images due to a single-pulse exposure, which made it possible to prevent the radiation damage from limiting the resolution. However, for practical reasons, the most suitable compact laser systems, usually Nd:YAG, which are commercially available produce nanosecond laser pulses with energies up to a few Joules with high repetition.
The use of solid targets is the most popular in SXCM systems. However, they are characterized by the problem of target debris, produced as a result of the laser ablation of the target material, which can cause sample damage when exposed to X-rays. To reduce the debris grazing incidences, X-ray optics in the form of a toroidal mirror was used to focus soft X-rays on a sample[15]. The debris problem was completely solved by applying laser-plasma soft X-ray sources based on a gas puff target in SXCM experiments[16].
3. Recording Media
The most common recording medium for SXCM is an X-ray resist in the form of thin (typically a few hundredths of a nanometer) layers of a photosensitive polymer on top of a silicon wafer or a glass slide. Depending on the type of photoresist, the radiation damages the molecular structure of the resist, decreasing the local molecular weight of the polymer chains, and making the resist susceptible to the chemical development procedure (positive photoresist, i.e., PMMA (poly(methyl methacrylate))[17].
Photoresists must ensure high resolution, preserve the cellular structure, and be suitable for cell imaging, ensuring cytocompatibility. Cells must strongly adhere to the resist. However, the most commonly used photoresist for biological applications is PMMA. Many resists are used in photolithography (e.g., DNQ-Novolac photoresist, epoxy-based polymer, hydrogen silsesquioxane) but are not suitable for cell imaging. Epoxidized Novolac resist (EPR) was also tested in SXCM, but without success[16].
Photoresists are “permanent” media, meaning that once recorded, the image of the object remains there for an extended period and can be digitized long after the exposure if the resist is properly preserved. This also means that the resist does not allow a refresh rate when trying to record changes in a sample. Resists require chemical development procedures but provide the highest spatial resolutions, which are mainly limited by digitization and equipment, such as SEM or AFM.
Another approach is to use lithium fluoride crystals (LiF) as the recording medium. Different centers are produced in LiF crystals when they are exposed to ionizing radiation[18]. Their emission intensity changes locally and is proportional to their density, thus revealing the 2-D X-ray intensity map and the radiographic image of the biological sample, which is in contact with LiF crystal during the exposure to X-rays. The drawback of this recording medium is the need for sufficiently high X-ray fluence values for color formation to obtain high-quality (i.e., high resolution) SXCM images. The advantage is that the LiF does not require a development procedure; however, the digitization is carried out with a fluorescence microscope, which is much more limited by its spatial resolution than SEM or AFM microscopes.
4. Exposure Environment
Most of the early SXCM experiments were performed by placing dehydrated specimens directly inside the vacuum chamber of the microscope, in which the soft X-rays propagated from the source to the specimen. With a resist such as PMMA as the recording medium, a relief structure of the sample was obtained and digitized with AFM. Because the primary goal of SXCM is to acquire high-resolution images of living specimens, the environment of the biological sample must be designed to maintain it in a hydrated state during exposure. For observation of living cells, soft X-rays are typically transmitted through silicon nitride membrane with thickness of 100–200 nm, covering the biological specimens cultivated directly onto the top surface of the photoresist. The whole assembly, consisting of the membrane, specimen in the culture medium, and the photoresist, are placed inside the vacuum chamber and sealed, employing specially designed mechanical capsules[19][20] to prevent evaporation and specimen drying.
Another approach is to place the assembly inside the pressurized environment chamber filled with helium[21] separated from the source chamber’s vacuum environment to prevent specimen drying. In this case, no particular effort is required to perform sealing, and the dismounting of the sample is much easier.
5. Development Procedure
The development of photoresists, the recording mediums in SXCM, has received considerable attention owing to their use in lithographic techniques. The development procedure is a critical step in SXCM because it affects image contrast and resolution. Irradiation of photoresists with soft X-rays results in bond breakage in the polymer structure, changing the molecular weights of resists locally and enabling these areas to be dissolved away in a chemical bath by solvents, such as methyl isobutyl ketone (MIBK), with the addition of isopropanol (IPA). Generally, each photoresist requires chemical solvents to dilute the irradiated area of the resist. The developer concentration and time exposure during the development process must be optimized experimentally[17]. Usually, a mixture of MIBK and IPA 1:1 (v:v) up to 1:3 (v:v) is used. A 1:1 (v/v) solution[22][20] means that the developer is chemically aggressive. A high concentration of methyl isobutyl ketone in the isopropanol increases the developing sensitivity but might blur some delicate structures in the photoresist. A more chemically gentle mixture of MIBK:IPA 1:3 (v:v) was recommended[21][23], as it provides sufficient sensitivity while preserving the nanometer height in delicate structures and features.
The resist is usually spin-coated on top of a silicon wafer, forming a uniform layer, typically 200–500 nm thick. The thickness is then tested by scratching the layer and measuring the height step using the AFM microscope. Moreover, after the resist is deposited, it must be calibrated for various SXR doses. The dose is changed by adjusting the number of SXR pulses used to expose the resist. For each dose, the depth of the relief is measured under the same developing conditions. It is necessary to optimize the SXR dose delivered to the resist to adjust its response and, finally, the dynamic range of the image contrast[22][20][24][25].
6. Digitization
The digitization process converts information stored in the recording medium, that is, the height information (relief pattern) in the photoresist or the density and transmittance of the color centers in the LiF crystal, into a digital image of the imprint, which is a representation of the specimen obtained during the recording phase. Historically, SEM and TEM were the techniques of choice for the digitization process. While SEM[26][7] and TEM[27][28] provided SXCM images with superior spatial resolution, the electron beam acting upon the photoresist sample causes the same effect as the soft X-ray radiation during the exposure/recording phase. This means that the e-beam during SEM/TEM image acquisition damages the photoresist and affects the information stored within it. For this reason, the AFM technique[29][18][30], which uses a sharp tip and van der Waals interactions between the end of the tip and the surface of the recording medium, provides much better height resolution and does not affect the sample in the manner of e-beam. LiF crystals require FM to digitize the information stored in the recording medium[30][31].
7. A Note on Spatial Resolution
It is not straightforward to assign the limit on spatial resolution in contact microscopy; however, several factors influence the spatial resolution, as seen in the following equation. This is an approximate relation, assuming all contributions have Gaussian profiles[2].
The δs is a factor related to shadowing (both wavelength-related diffraction and geometrically related to variable sample–resist distances due to non-zero thickness of the sample), δphr is a factor related to the spatial resolution of the photoresist, e.g., for HSQ/PMMA combinations below 10 nm[4], PMMA ~10 nm[32], and δtip is a factor related to the diameter of the AFM probe used to perform photoresist scanning after the exposure, typically 20 nm in diameter, but can be as sharp as ~2 nm for carbon nanotube probes[6]. Taking into account all the above factors, the spatial resolution in SXCM is routinely much better than 100 nm[8][33], allowing a practical limit of ~10 nm[28][34].
More detailed numerical studies were also performed[35], in which the effects of sample and resist absorption and diffraction were considered, as was the process of isotropic development of the photoresist. The SXCM image resolution heavily depends on the exposure and the sample-to-resist distance. The contrast of small features imprinted in the photoresist depends significantly on the development procedure. If researchers are not careful, the information on the smallest features may be destroyed by excessively aggressive or long development. This is why these issues must be kept in mind during the postprocessing and interpretation of the SXCM images of high-resolution and low-contrast objects, such as biological structures.
8. Desk-Top Laboratory SXCM System
Almost all research on SXCM to date has been carried out using laboratory systems that are not adapted to use by biologists in their laboratory practice. In many articles, the authors emphasize the potential of this simple imaging technique and its usefulness in studying the structures of cells; however, so far, no attempt has been made to commercialize the SXCM microscopy system. The conceptual design of the laboratory microscope was presented only in[36]. Here, one group present the compact laboratory system for SXCM microscopy[21][37], which is believed to meet these expectations.
The schematic of the system is presented in Figure 2. The biological sample is placed on the resist in the form of a 500-nanometer-thick PMMA film spun on a silicon wafer. The plate is attached to a holder mounted inside a helium-filled chamber. The design of the holder and its chamber allows the sample to be positioned at a distance of 15 mm from the source of soft X-rays. The source is a laser plasma resulting from the interaction of laser pulses with a double-stream gas puff target. Laser pulses with a duration of 4 ns and energy up to 0.8 J are generated with a repetition frequency of 10 Hz in a commercially available Nd: YAG laser system (EXPLA). The target is formed by pulsed injection of a small amount of argon gas under high pressure into an annular stream of helium surrounding the argon gas stream using a specially designed electromagnetic valve system. The use of the argon gas puff target allows the production of soft X-ray radiation with high efficiency, comparable to a solid target, without the harmful effect of the target debris. The valve producing the gas puff target is mounted inside the vacuum source chamber in such a way that the target is formed at the focus of the laser beam produced by the aspherical focusing lens. The holder chamber is equipped with a 100-nanometer-thick Si3N4 membrane, which separates the chamber’s helium atmosphere from the vacuum inside the source chamber. This allows the sample to be placed near the source without venting the vacuum chamber. Additionally, a 200-nanometer-thick Ti membrane is used to eliminate visible light from the plasma. The source produces soft X-ray radiation in the “water window” range, with a maximum wavelength of about 3 nm amd energy fluence of about 5 μJ/cm2/pulse, corresponding to the photon fluence of about 103 photons/μm2/pulse. This fluence makes it possible to obtain the SXCM image of a sample as a result of 100–200 laser pulses (10–20 s exposition time). The whole system can be mounted on an optical table with dimensions of approximately 1 × 1 m2. Detailed information on the presented system and the results of preliminary studies on SXCM can be found in the previous publications[38][21].
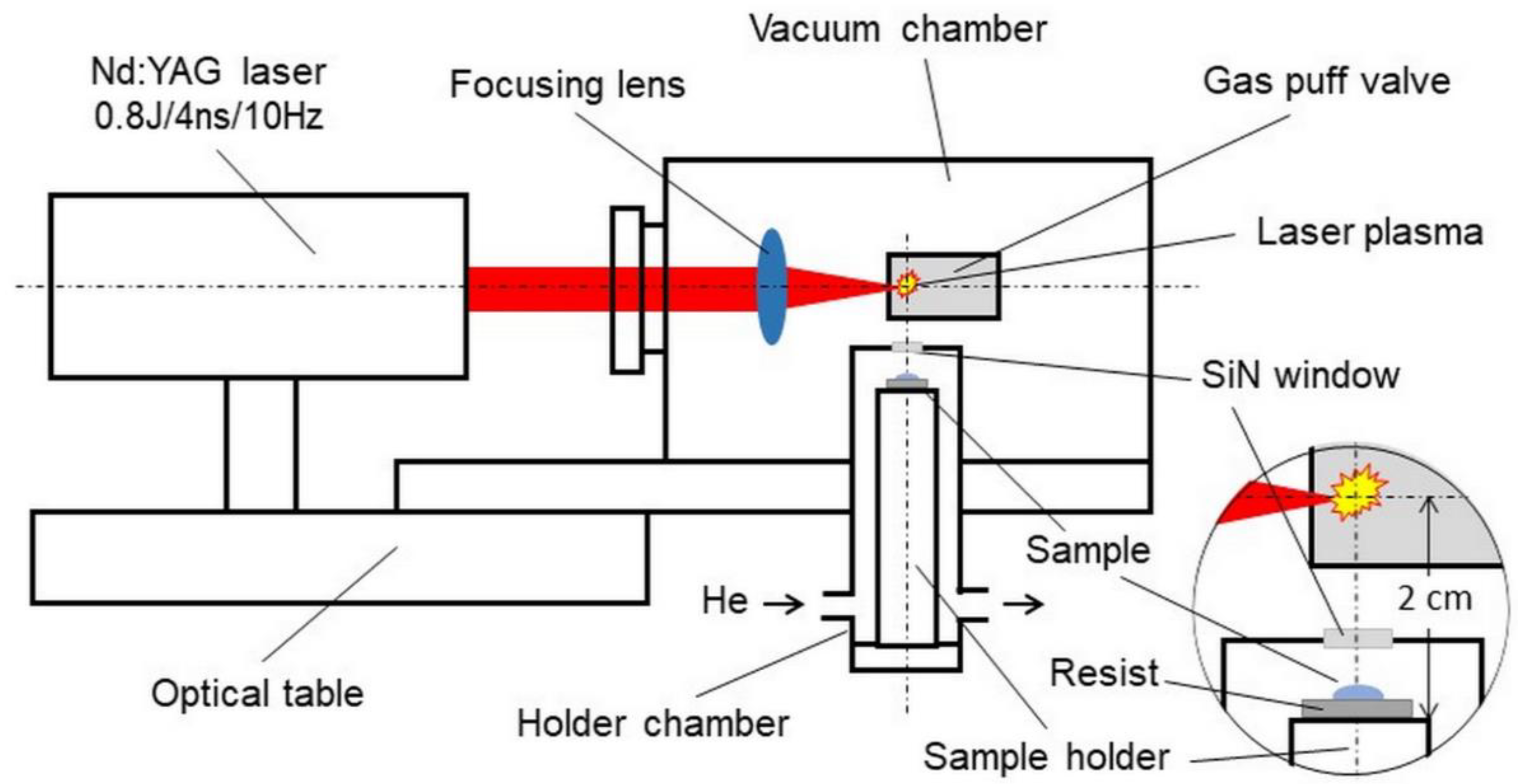
Figure 2. Schematic of the desk-top laboratory system for soft X-ray contact microscopy (SXCM) based on a laser plasma soft X-ray source with a double-stream gas puff target.
References
- V. E. Cosslett; Microscopy with X-Rays. Nature 1959, 183, 1423-1427, 10.1038/1831423a0.
- Wachulak, Przemysław W.. 21st Century Nanoscience – A Handbook; Sattler, Klaus D. , Eds.; CRC Press: Boca Raton, 2020; pp. chapter 7.
- R.C. Greulich; A. Engström; A new approach to high resolution microradiography using extremely soft X-rays. Experimental Cell Research 1956, 10, 251-254, 10.1016/0014-4827(56)90097-0.
- Marcus Rommel; Bengt Nilsson; Piotr Jedrasik; Valentina Bonanni; Alexandre Dmitriev; Jürgen Weis; Sub-10nm resolution after lift-off using HSQ/PMMA double layer resist. Microelectronic Engineering 2013, 110, 123-125, 10.1016/j.mee.2013.02.101.
- Ralph Feder; David Sayre; Eberhard Spiller; John Topalian; Janos Kirz; Specimen replication for electron microscopy using x rays and x‐ray resist. Journal of Applied Physics 1976, 47, 1192-1193, 10.1063/1.322704.
- Ramsey M. Stevens; New carbon nanotube AFM probe technology. Materials Today 2009, 12, 42-45, 10.1016/s1369-7021(09)70276-7.
- E. Spiller; R. Feder; J. Topalian; D. Eastman; W. Gudat; D. Sayre; X-Ray Microscopy of Biological Objects with Carbon K α and with Synchrotron Radiation. Science 1976, 191, 1172-1174, 10.1126/science.1257741.
- M Kado; M Kishimoto; S Tamotsu; K Yasuda; K Shinohara; In situ observation of cellular organelles with a contact x-ray microscope. Journal of Physics: Conference Series 2013, 463, 012056, 10.1088/1742-6596/463/1/012056.
- R. Feder; E. Spiller; J. Topalian; A. N. Broers; W. Gudat; B. J. Panessa; Z. A. Zadunaisky; J. Sedat; High-Resolution Soft X-Ray Microscopy. Science 1977, 197, 259-260, 10.1126/science.406670.
- T.W. Ford; A.D. Stead; R.A. Cotton; Soft X-ray contact microscopy of biological materials. Electron Microscopy Reviews 1991, 4, 269-292, 10.1016/0892-0354(91)90006-x.
- R. J. Rosser; R. Feder; A. Ng; F. Adams; P. Celliers; R. J. Speer; Nondestructive single-shot soft x-ray lithography and contact microscopy using a laser-produced plasma source. Applied Optics 1987, 26, 4313-4318, 10.1364/ao.26.004313.
- Masataka Kado; Martin C. Richardson; Kai Gaebel; David Scott Torres; Jayshree Rajyaguru; Michael J. Muszynski; Ultrastructural imaging and molecular modeling of live bacteria using soft x-ray contact microscopy with nanoseconds laser-plasma radiation. SPIE's 1995 International Symposium on Optical Science, Engineering, and Instrumentation 1995, 2523, 194-202, 10.1117/12.220979.
- A G Michette; P C Cheng; R W Easons; R Feder; F O'neill; Y Owadano; R J Rosser; P Rumsby; M J Shaw; Soft X-ray contact microscopy using laser plasma sources. Journal of Physics D: Applied Physics 1986, 19, 363-372, 10.1088/0022-3727/19/3/008.
- P. Albertano; L. Reale; L. Palladino; A. Reale; R. Cotton; S. Bollanti; P. DI Lazzaro; F. Flora; N. Lisi; A. Nottola; et al.K. Vigli PapadakiT. LetardiD. BanaiA. ContiM. MoretA. Grilli X‐ray contact microscopy using an excimer laser plasma source with different target materials and laser pulse durations. Journal of Microscopy 1997, 187, 96-103, 10.1046/j.1365-2818.1997.2120768.x.
- R. J. Rosser; R. Feder; A. Ng; F. Adams; M. Caldarolo; P. Celliers; P. C. Cheng; L. Silva; D. Parfeniuk; R. J. Speer; et al. Biological specimens imaged by soft x-ray contact microscopy using a plasma source produced with a laboratory sized laser. Journal of Microscopy 1986, 144, RP5-RP6, 10.1111/j.1365-2818.1986.tb02792.x.
- Y. Kinjo; M. Watanabe; H. Fiedorowicz; H. Daido; E. Yanase; S. Fujii; E. Sato; K. Shinohara; Fine structures of human chromosomes observed by X-ray contact microscopy coupled with atomic force microscopy. Journal de Physique IV (Proceedings) 2003, 104, 313-316, 10.1051/jp4:200300088.
- Shazia Yasin; D.G. Hasko; H. Ahmed; Comparison of MIBK/IPA and water/IPA as PMMA developers for electron beam nanolithography. Microelectronic Engineering 2002, 61-62, 745-753, 10.1016/s0167-9317(02)00468-9.
- Kinjo, Y., Watanabe, M., Ito, A., & Shinohara, K. (2005). X-ray Microscopy and Chromosome Research. In 8th International Conference on X-ray Microscopy; IPAP Conference Series (Vol. 7, pp. 227-229).
- D. Batani; C. Botto; M. Moret; M. Milani; G. Lucchini; K. Eidmann; F. Cotelli; C. Lora Lamia Donin; G. Poletti; T. Ford; et al.A. Stead The use of high energy laser-plasma sources in soft X-ray contact microscopy of living biological samples. The European Physical Journal D 2002, 21, 167-179, 10.1140/epjd/e2002-00194-y.
- Toshikazu Majima; Soft X-ray imaging of living cells in water: flash contact soft X-ray microscope. TrAC Trends in Analytical Chemistry 2004, 23, 520-526, 10.1016/s0165-9936(04)00735-6.
- M.G. Ayele; P.W. Wachulak; J. Czwartos; D. Adjei; A. Bartnik; Ł. Wegrzynski; M. Szczurek; L. Pina; H. Fiedorowicz; Development and characterization of a laser-plasma soft X-ray source for contact microscopy. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 2017, 411, 35-43, 10.1016/j.nimb.2017.03.082.
- A.C Cefalas; E Sarantopoulou; Z Kollia; P Argitis; E Tegou; T.W Ford; A.D Stead; C.N Danson; D Neely; S Kobe; et al. Nanostructured imaging of biological specimens in vivo with laser plasma X-ray contact microscopy. Materials Science and Engineering: C 2003, 23, 105-108, 10.1016/s0928-4931(02)00242-4.
- Y. Wang; Chris Jacobsen; A numerical study of resolution and contrast in soft X‐ray contact microscopy. Journal of Microscopy 1998, 191, 159-169, 10.1046/j.1365-2818.1998.00353.x.
- Y. Wang; Chris Jacobsen; A numerical study of resolution and contrast in soft X‐ray contact microscopy. Journal of Microscopy 1998, 191, 159-169, 10.1046/j.1365-2818.1998.00353.x.
- M. Kado; M. Ishino; S. Tamotsu; K. Yasuda; M. Kishimoto; M. Nishikino; Y. Kinjo; K. Shinohara; Ian McNulty; Catherine Eyberger; et al.Barry Lai Observation of Organelles in Leydig Cells by Contact Soft X-Ray Microscopy with a Laser Plasma X-Ray Source. AIP Conference Proceedings 2011, 1365, 391, 10.1063/1.3625385.
- Barbara J. Panessa-Warren; Contact microscopy with synchrotron radiation. Biological Trace Element Research 1987, 12, 167-183, 10.1007/bf02796678.
- Cheng, P. C.; Shinozaki, D. M.; Tan, K. H.. Recent Advances in Contact Imaging of Biological Materials; Cheng, Ping-chin Jan, Gwo-jen, Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 1987; pp. 65-104.
- Ralph Feder; Jonathan L. Costa; Praveen Chaudhari; David Sayre; Improved Detail in Biological Soft X-ray Microscopy: Study of Blood Platelets. Science 1981, 212, 1398-1400, 10.1126/science.7233227.
- Yoshimasa Yamamoto; Kunio Shinohara; Application of X-ray microscopy in analysis of living hydrated cells. The Anatomical Record 2002, 269, 217-223, 10.1002/ar.10166.
- L. Reale; F. Bonfigli; A. Lai; F. Flora; P. Albertano; M. L. DI Giorgio; L. Mezi; R.M. Montereali; A. Faenov; T. Pikuz; et al.S. AlmavivaM. FrancucciP. GaudioS. MartellucciM. RichettaA. Poma Contact X-ray microscopy of living cells by using LiF crystal as imaging detector. Journal of Microscopy 2015, 258, 127-139, 10.1111/jmi.12226.
- G. Baldacchini; S. Bollanti; F. Bonfigli; F. Flora; P. Di Lazzaro; A. Lai; T. Marolo; R. M. Montereali; D. Murra; A. Faenov; et al.T. PikuzE. NichelattiG. TomassettiA. RealeL. RealeA. RitucciT. LimongiL. PalladinoM. FrancucciS. MartellucciG. Petrocelli Soft x-ray submicron imaging detector based on point defects in LiF. Review of Scientific Instruments 2005, 76, 113104, 10.1063/1.2130930.
- Wenchuang (Walter) Hu; Koshala Sarveswaran; Marya Lieberman; Gary H. Bernstein; Sub-10 nm electron beam lithography using cold development of poly(methylmethacrylate). Journal of Vacuum Science & Technology B: Microelectronics and Nanometer Structures 2004, 22, 1711, 10.1116/1.1763897.
- M.G. Ayele; J. Czwartos; D. Adjei; P. Wachulak; I.U. Ahad; A. Bartnik; Ł. Wegrzynski; M. Szczurek; R. Jarocki; H. Fiedorowicz; et al.M. LekkaK. PogodaJ. Gostek Contact Microscopy using a Compact Laser Produced Plasma Soft X-Ray Source. Acta Physica Polonica A 2016, 129, 237-240, 10.12693/aphyspola.129.237.
- L. Beese; R. Feder; D. Sayre; Contact x-ray microscopy. A new technique for imaging cellular fine structure. Biophysical Journal 1986, 49, 259-268, 10.1016/s0006-3495(86)83639-6.
- Y. Wang; Chris Jacobsen; A numerical study of resolution and contrast in soft X‐ray contact microscopy. Journal of Microscopy 1998, 191, 159-169, 10.1046/j.1365-2818.1998.00353.x.
- Martin C. Richardson; Kunio Shinohara; Kazuo A. Tanaka; Yasuhito Kinjo; Naomi Ikeda; Masataka Kado; Pulsed x-ray microscopy of biological specimens with laser plasma sources. Soft X-Ray Microscopy 1993, 1741, 133-142, 10.1117/12.138727.
- Paulina Osuchowska; Przemysław Wachulak; Agata Nowak-Stępniowska; Andrzej Bartnik; Kajangi Gnanachandran; Małgorzata Lekka; Joanna Czwartos; Henryk Fiedorowicz; Elżbieta Trafny; Imaging of Cell Structures Using Optimized Soft X-ray Contact Microscopy. Applied Sciences 2020, 10, 6895, 10.3390/app10196895.
- Paulina Osuchowska; Przemysław Wachulak; Wiktoria Kasprzycka; Agata Nowak-Stępniowska; Maciej Wakuła; Andrzej Bartnik; Henryk Fiedorowicz; Elżbieta Trafny; Adhesion of Triple-Negative Breast Cancer Cells under Fluorescent and Soft X-ray Contact Microscopy. International Journal of Molecular Sciences 2021, 22, 7279, 10.3390/ijms22147279.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
21 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




