
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stefan Hobi | -- | 1816 | 2022-07-20 04:37:42 |
Video Upload Options
Malassezia spp. are commensals of the skin, oral/sinonasal cavity, lower respiratory and gastrointestinal tract. Eighteen species have been recovered from humans, other mammals and birds. They can also be isolated from diverse environments, suggesting an evolutionary trajectory of adaption from an ecological niche in plants and soil to the mucocutaneous ecosystem of warm-blooded vertebrates.
1. Introduction
2. Classification of Malassezia Yeasts
|
Species |
Reference Strain/GenBank Accession Genome Number |
Described Hosts |
Clade |
|---|---|---|---|
|
M. furfur |
CBS 14141, GCA_009938135 |
Human, Cat, Dog, Cattle, Pig, Goat, Elk, Horse, Sheep, Elephant, Monkey, Ostrich, Pelican |
A |
|
M. brasiliensis * |
MA 1455 |
Parrot |
A |
|
M. yamatoensis |
MY9725, GCA_001264885 |
Human, Cat |
A |
|
M. psittaci * |
MA 1454 |
Parrot |
A |
|
M. obtusa |
CBS 7876, GCA_001264985 |
Human, Cat, Dog, Goat, Horse |
A |
|
M. japonica |
CBS 9431, GCA_001264785 |
Human, Cat |
A |
|
M. vespertilionis |
CBS 15041, GCA_002818225 |
Bat |
A |
|
M. globosa |
CBS 7966, GCA_001264805 |
Human, Cat, Dog, Cattle, Goat, Horse, Sheep, Cheetah |
B |
|
M. restricta |
CBS 7877, GCA_001264765 |
Human, Cat, Dog, Cattle, Goat, Horse, Sheep |
B |
|
M. arunalokei |
CBS 13387, GCA_020085095 |
Human, Dog |
B |
|
M. sympodialis |
ATCC 42132, GCA_001264925 |
Human, Dog, Cat, Pig, Cattle, Goat, Horse, Sheep, Chicken |
B |
|
M. dermatis |
CBS 9169, GCA_001264665 |
Human, Cat |
B |
|
M. caprae |
CBS 10434, GCA_001264625 |
Goat, Horse, Human |
B |
|
M. equina |
CBS 9969, GCA_001264685 |
Horse, Cattle |
B |
|
M. nana |
JCM 12085, GCA_001600835 |
Cat, Dog, Cattle, Horse |
B |
|
M. pachydermatis |
CBS 1879, GCA_001264975 |
Human, Dog, Cat, Pig, Goat, Rabbit, Various exotic and wild mammals, Birds (Thraupidae, Macaw) |
B |
|
M. cuniculi |
CBS 11721, GCA_001264635 |
Rabbit |
C |
|
M. slooffiae |
CBS 7956, GCA_001264965 |
Human, Cat Cattle, Sheep, Pig, Goat, Horse |
C |
3. Malassezia Species in the Environment and Possible Vectors
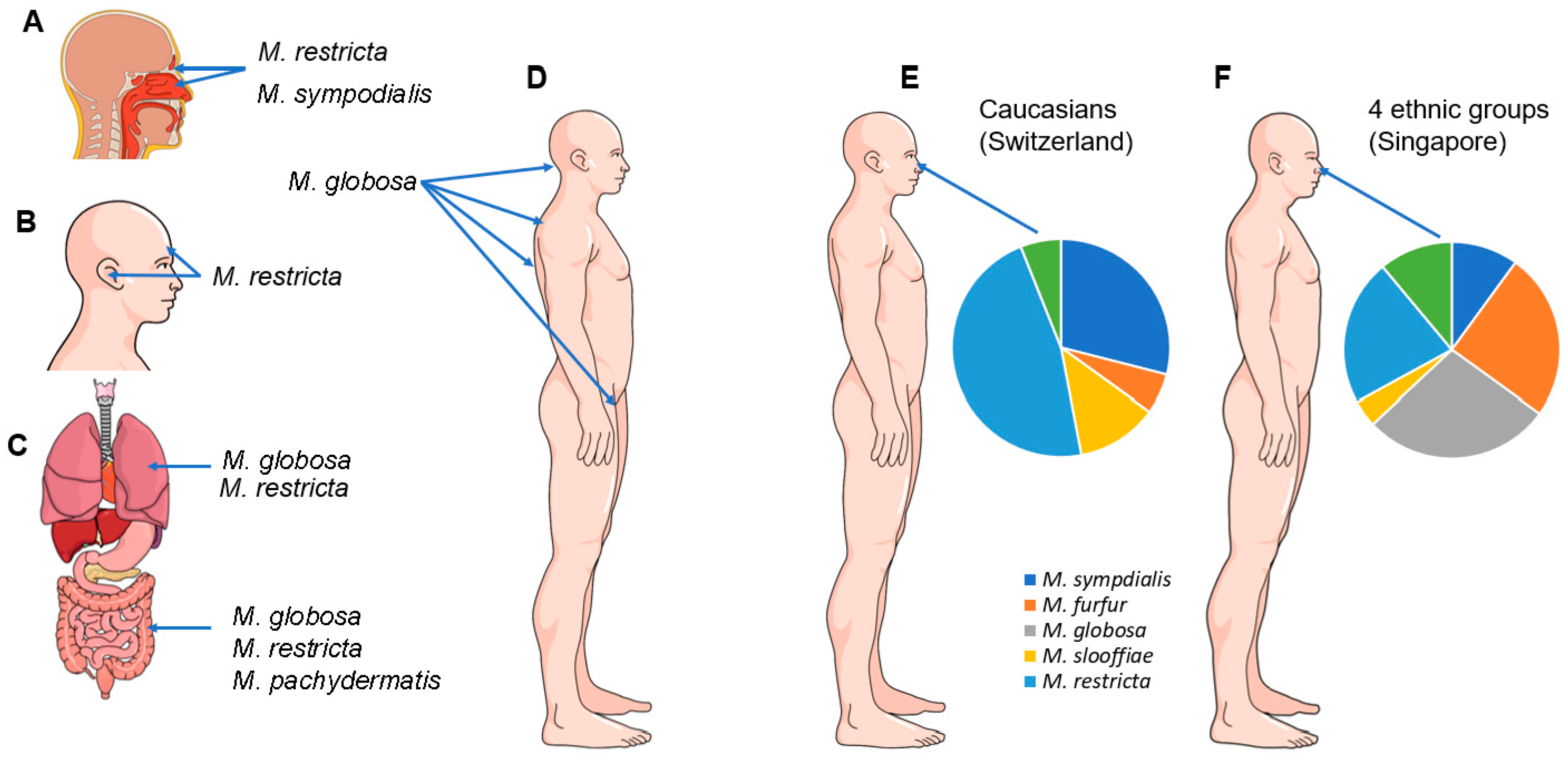
4. Malassezia Species and Their Role as Commensals in Humans
References
- Wu, G.; Zhao, H.; Li, C.; Rajapakse, M.P.; Wong, W.C.; Xu, J.; Saunders, C.W.; Reeder, N.L.; Reilman, R.A.; Scheynius, A.; et al. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet. 2015, 11, e1005614.
- Bond, R.; Morris, D.O.; Guillot, J.; Bensignor, E.J.; Robson, D.; Mason, K.V.; Kano, R.; Hill, P.B. Biology, diagnosis and treatment of Malassezia dermatitis in dogs and cats Clinical Consensus Guidelines of the World Association for Veterinary Dermatology. Vet. Dermatol. 2020, 31, 28–74.
- Saadatzadeh, M.R. The Immunology of the Mycelial Phase of Malassezia; University of Leeds: Leeds, UK, 1998.
- Saadatzadeh, M.; Ashbee, H.; Holland, K.; Ingham, E. Production of the mycelial phase of Malassezia in vitro. Sabouraudia 2001, 39, 487–493.
- Saadatzadeh, M.; Ashbee, H.; Cunliffe, W.; Ingham, E. Cell-mediated immunity to the mycelial phase of Malassezia spp. in patients with pityriasis versicolor and controls. Br. J. Dermatol. 2001, 144, 77–84.
- Prohic, A.; Jovovic Sadikovic, T.; Krupalija-Fazlic, M.; Kuskunovic-Vlahovljak, S. Malassezia species in healthy skin and in dermatological conditions. Int. J. Dermatol. 2016, 55, 494–504.
- Gioti, A.; Nystedt, B.R.; Li, W.; Xu, J.; Andersson, A.; Averette, A.F.; MŘnch, K.; Wang, X.; Kappauf, C.; Kingsbury, J.M. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. MBio 2013, 4, e00572-12.
- Shifrine, M.; Marr, A. The requirement of fatty acids by Pityrosporum ovale. Microbiology 1963, 32, 263–270.
- Brunke, S.; Hube, B. MfLIP1, a gene encoding an extracellular lipase of the lipid-dependent fungus Malassezia furfur. Microbiology 2006, 152, 547–554.
- Juntachai, W.; Oura, T.; Murayama, S.Y.; Kajiwara, S. The lipolytic enzymes activities of Malassezia species. Sabouraudia 2009, 47, 477–484.
- Puig, L.; Bragulat, M.R.; Castella, G.; Cabanes, F.J. Characterization of the species Malassezia pachydermatis and re-evaluation of its lipid dependence using a synthetic agar medium. PLoS ONE 2017, 12, e0179148.
- Vest, B.E.; Krauland, K. Malassezia Furfur. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020.
- Saunte, D.M.L.; Gaitanis, G.; Hay, R.J. Malassezia-Associated Skin Diseases, the Use of Diagnostics and Treatment. Front. Cell. Infect. Microbiol. 2020, 10, 112.
- Rhimi, W.; Theelen, B.; Boekhout, T.; Otranto, D.; Cafarchia, C. Malassezia spp. Yeasts of Emerging Concern in Fungemia. Front. Cell Infect. Microbiol. 2020, 10, 370.
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151.
- Iatta, R.; Cafarchia, C.; Cuna, T.; Montagna, O.; Laforgia, N.; Gentile, O.; Rizzo, A.; Boekhout, T.; Otranto, D.; Montagna, M.T. Bloodstream infections by Malassezia and Candida species in critical care patients. Med. Mycol. 2014, 52, 264–269.
- Ilahi, A.; Hadrich, I.; Neji, S.; Trabelsi, H.; Makni, F.; Ayadi, A. Real-Time PCR Identification of Six Malassezia Species. Curr. Microbiol. 2017, 74, 671–677.
- Pedrosa, A.F.; Lisboa, C.; Rodrigues, A.G. Malassezia infections with systemic involvement: Figures and facts. J. Dermatol. 2018, 45, 1278–1282.
- Salkin, I.; Stone, W.; Gordon, M. Association of Malassezia (Pityrosporum) pachydermatis with sarcoptic mange in New York State. J. Wildl. Dis. 1980, 16, 509–514.
- Breuer-Strosberg, R.; Hochleithner, M.; Kuttin, E. Malassezia pachydermatis isolation from a scarlet macaw. Mycoses 1990, 33, 247–250.
- Guillot, J.; Petit, T.; Degorce-Rubiales, F.; Guého, E.; Chermette, R. Dermatitis caused by Malassezia pachydermatis in a California sea lion (Zalophus californianus). Vet. Rec. 1998, 142, 311–312.
- Nakagaki, K.; Hata, K.; Iwata, E.; Takeo, K. Malassezia pachydermatis isolated from a South American sea lion (Otaria byronia) with dermatitis. J. Vet. Med. Sci. 2000, 62, 901–903.
- Begerow, D.; Bauer, R.; Boekhout, T. Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol. Res. 2000, 104, 53–60.
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371.
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547.
- Wang, Q.-M.; Theelen, B.; Groenewald, M.; Bai, F.-Y.; Boekhout, T. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Pers. Mol. Phylogeny Evol. Fungi 2014, 33, 41.
- Lorch, J.M.; Palmer, J.M.; Vanderwolf, K.J.; Schmidt, K.Z.; Verant, M.L.; Weller, T.J.; Blehert, D.S. Malassezia vespertilionis sp. nov.: A new cold-tolerant species of yeast isolated from bats. Persoonia 2018, 41, 56–70.
- Castella, G.; Coutinho, S.D.; Cabanes, F.J. Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med. Mycol. 2014, 52, 99–105.
- Cabanes, F.J.; Theelen, B.; Castella, G.; Boekhout, T. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res. 2007, 7, 1064–1076.
- Cabañes, F.; Vega, S.; Castellá, G. Malassezia cuniculi sp. nov. a novel yeast species isolated from rabbit skin. Med. Mycol. 2011, 49, 40–48.
- Cabanes, F.J.; Coutinho, S.D.; Puig, L.; Bragulat, M.R.; Castella, G. New lipid-dependent Malassezia species from parrots. Rev. Iberoam. Micol. 2016, 33, 92–99.
- Park, M.; Cho, Y.J.; Lee, Y.W.; Jung, W.H. Whole genome sequencing analysis of the cutaneous pathogenic yeast Malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff. Mycoses 2017, 60, 188–197.
- Morand, S.C.; Bertignac, M.; Iltis, A.; Kolder, I.; Pirovano, W.; Jourdain, R.; Clavaud, C. Complete Genome Sequence of Malassezia restricta CBS 7877, an Opportunist Pathogen Involved in Dandruff and Seborrheic Dermatitis. Microbiol. Resour. Announc. 2019, 8, e01543-18.
- Chow, N.A.; Chinn, R.; Pong, A.; Schultz, K.; Kim, J.; Gade, L.; Jackson, B.R.; Beer, K.D.; Litvintseva, A.P. Use of whole-genome sequencing to detect an outbreak of Malassezia pachydermatis infection and colonization in a neonatal intensive care unit—California, 2015–2016. Infect. Control. Hosp. Epidemiol. 2020, 41, 851–853.
- D’Andreano, S.; Viñes, J.; Francino, O. Whole-Genome Sequencing and De Novo Assembly of Malassezia pachydermatis Isolated from the Ear Canal of a Dog with Otitis. Microbiol. Resour. Announc. 2021, 10, e00205-21.
- Amend, A. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog. 2014, 10, e1004277.
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.; Heitman, J.; Hom, E.F.; Ianiri, G.; Jones, A.C. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10, e01189-18.
- Theelen, B.; Cafarchia, C.; Gaitanis, G.; Bassukas, I.D.; Boekhout, T.; Dawson, T.L., Jr. Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 2018, 56 (Suppl. S1), S10–S25.
- Arenz, B.E.; Held, B.W.; Jurgens, J.A.; Farrell, R.L.; Blanchette, R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 2006, 38, 3057–3064.
- Fell, J.W.; Scorzetti, G.; Connell, L.; Craig, S. Biodiversity of micro-eukaryotes in Antarctic Dry Valley soils with<5% soil moisture. Soil Biol. Biochem. 2006, 38, 3107–3119.
- Bass, D.; Howe, A.; Brown, N.; Barton, H.; Demidova, M.; Michelle, H.; Li, L.; Sanders, H.; Watkinson, S.C.; Willcock, S.; et al. Yeast forms dominate fungal diversity in the deep oceans. Proc. Biol. Sci. 2007, 274, 3069–3077.
- Lai, X.; Cao, L.; Tan, H.; Fang, S.; Huang, Y.; Zhou, S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007, 1, 756–762.
- Le Calvez, T.; Burgaud, G.; Mahe, S.; Barbier, G.; Vandenkoornhuyse, P. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 2009, 75, 6415–6421.
- Gao, Z.; Johnson, Z.I.; Wang, G. Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. ISME J. 2010, 4, 111–120.
- Roy, M.; Watthana, S.; Stier, A.; Richard, F.; Vessabutr, S.; Selosse, M.A. Two mycoheterotrophic orchids from Thailand tropical dipterocarpacean forests associate with a broad diversity of ectomycorrhizal fungi. BMC Biol. 2009, 7, 51.
- Jebaraj, C.S.; Raghukumar, C.; Behnke, A.; Stoeck, T. Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol. Ecol. 2010, 71, 399–412.
- Edgcomb, V.P.; Beaudoin, D.; Gast, R.; Biddle, J.F.; Teske, A. Marine subsurface eukaryotes: The fungal majority. Environ. Microbiol. 2011, 13, 172–183.
- Singh, P.; Raghukumar, C.; Verma, P.; Shouche, Y. Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microb. Ecol. 2011, 61, 507–517.
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522.
- Orsi, W.; Biddle, J.F.; Edgcomb, V. Deep sequencing of subseafloor eukaryotic rRNA reveals active Fungi across marine subsurface provinces. PLoS ONE 2013, 8, e56335.
- Zhang, T.; Wang, N.F.; Zhang, Y.Q.; Liu, H.Y.; Yu, L.Y. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Sci. Rep. 2015, 5, 14524.
- Pang, K.L.; Guo, S.Y.; Chen, I.A.; Burgaud, G.; Luo, Z.H.; Dahms, H.U.; Hwang, J.S.; Lin, Y.L.; Huang, J.S.; Ho, T.W.; et al. Insights into fungal diversity of a shallow-water hydrothermal vent field at Kueishan Island, Taiwan by culture-based and metabarcoding analyses. PLoS ONE 2019, 14, e0226616.
- Gao, Z.; Li, B.; Zheng, C.; Wang, G. Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl. Environ. Microbiol. 2008, 74, 6091–6101.
- Amend, A.S.; Barshis, D.J.; Oliver, T.A. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J. 2012, 6, 1291–1301.
- Renker, C.; Alphei, J.; Buscot, F. Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol. Fertil. Soils 2003, 37, 70–72.
- Tondello, A.; Vendramin, E.; Villani, M.; Baldan, B.; Squartini, A. Fungi associated with the southern Eurasian orchid Spiranthes spiralis (L.) Chevall. Fungal Biol. 2012, 116, 543–549.
- Quandt, A.; Glasco, A.; James, T. Intestinal Mycobiome Variation Across Geography and Phylogeny in the Snail Genus Conus. Genetics Society of America. In Proceedings of the 29th Fungal Genetics Conference Asilomar, Asilomar, CA, USA, 14–19 March 2017; Volume 20.
- Malacrinò, A.; Schena, L.; Campolo, O.; Laudani, F.; Mosca, S.; Giunti, G.; Strano, C.P.; Palmeri, V. A metabarcoding survey on the fungal microbiota associated to the olive fruit fly. Microb. Ecol. 2017, 73, 677–684.
- Duarte, E.; Resende, J.; Rosa, C.; Hamdan, J. Prevalence of yeasts and mycelial fungi in bovine parasitic otitis in the State of Minas Gerais, Brazil. J. Vet. Med. Ser. B 2001, 48, 631–635.
- Elhady, A.; Gine, A.; Topalovic, O.; Jacquiod, S.; Sorensen, S.J.; Sorribas, F.J.; Heuer, H. Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS ONE 2017, 12, e0177145.
- Simmons, R.B.; Gueho, E. A new species of Malassezia. Mycol. Res. 1990, 94, 1146–1149.
- Nakabayashi, A.; Sei, Y.; Guillot, J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 2000, 38, 337–341.
- Bernier, V.; Weill, F.X.; Hirigoyen, V.; Elleau, C.; Feyler, A.; Labrèze, C.; Sarlangue, J.; Chène, G.; Couprie, B.; Taïeb, A. Skin colonization by Malassezia species in neonates: A prospective study and relationship with neonatal cephalic pustulosis. Arch. Dermatol. 2002, 138, 215–218.
- Sugita, T.; Takashima, M.; Shinoda, T.; Suto, H.; Unno, T.; Tsuboi, R.; Ogawa, H.; Nishikawa, A. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J. Clin. Microbiol. 2002, 40, 1363–1367.
- Sugita, T.; Takashima, M.; Kodama, M.; Tsuboi, R.; Nishikawa, A. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J. Clin. Microbiol. 2003, 41, 4695–4699.
- Sugita, T.; Tajima, M.; Takashima, M.; Amaya, M.; Saito, M.; Tsuboi, R.; Nishikawa, A. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol. Immunol. 2004, 48, 579–583.
- Gupta, A.K.; Boekhout, T.; Theelen, B.; Summerbell, R.; Batra, R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J. Clin. Microbiol. 2004, 42, 4253–4260.
- Batra, R.; Boekhout, T.; Guého, E.; Cabanes, F.J.; Dawson, T.L., Jr.; Gupta, A.K. Malassezia Baillon, emerging clinical yeasts. FEMS Yeast Res. 2005, 5, 1101–1113.
- Boekhout, T.; Guého-Kellermann, E.; Mayser, P.; Velegraki, A. Malassezia and the Skin: Science and Clinical Practice; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010.
- Gaitanis, G.; Velegraki, A.; Mayser, P.; Bassukas, I.D. Skin diseases associated with Malassezia yeasts: Facts and controversies. Clin. Dermatol. 2013, 31, 455–463.
- Cabañes, F. Malassezia Yeasts: How Many Species Infect Humans and Animals? PLoS Pathog. 2014, 10, e1003892.
- Aguirre, C.; Euliarte, C.; Finquelievich, J.; de los Ángeles Sosa, M.; Giusiano, G. Fungemia and interstitial lung compromise caused by Malassezia sympodialis in a pediatric patient. Rev. Iberoam. Micol. 2015, 32, 118–121.
- Honnavar, P.; Chakrabarti, A.; Dogra, S.; Handa, S.; Rudramurthy, S.M. Phenotypic and molecular characterization of Malassezia japonica isolated from psoriasis vulgaris patients. J. Med. Microbiol. 2015, 64, 232–236.
- Honnavar, P.; Prasad, G.S.; Ghosh, A.; Dogra, S.; Handa, S.; Rudramurthy, S.M. Malassezia arunalokei sp. nov. a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from India. J. Clin. Microbiol. 2016, 54, 1826–1834.
- Patron, R.L. A 34-year-old man with cough, lung nodules, fever, and eosinophilia. Clin. Infect. Dis. 2016, 63, 1525–1526.
- Niemiec, B.A.; Gawor, J.; Tang, S.; Prem, A.; Krumbeck, J.A. The mycobiome of the oral cavity in healthy dogs and dogs with periodontal disease. Am. J. Vet. Res. 2021, 83, 42–49.
- Nagata, R.; Nagano, H.; Ogishima, D.; Nakamura, Y.; Hiruma, M.; Sugita, T. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatrics Int. 2012, 54, 350–355.
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253.
- Jo, J.-H.; Deming, C.; Kennedy, E.A.; Conlan, S.; Polley, E.C.; Ng, W.-I.; Segre, J.A.; Kong, H.H.; Program, N.C.S. Diverse human skin fungal communities in children converge in adulthood. J. Investig. Dermatol. 2016, 136, 2356–2363.
- Ward, T.L.; Dominguez-Bello, M.G.; Heisel, T.; Al-Ghalith, G.; Knights, D.; Gale, C.A. Development of the human mycobiome over the first month of life and across body sites. MSystems 2018, 3, e00140-17.
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370.
- Gueho, E.; Midgley, G.; Guillot, J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek 1996, 69, 337–355.
- Aspiroz, C.; Moreno, L.-A.; Rezusta, A.; Rubio, C. Differentiation of three biotypes of Malassezia species on human normal skin. Correspondence with M. globosa, M. sympodialis and M. restricta. Mycopathologia 1999, 145, 69–74.
- Gupta, A.; Kohli, Y.; Summerbell, R.; Faergemann, J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 2001, 39, 243–251.
- Gemmer, C.M.; DeAngelis, Y.M.; Theelen, B.; Boekhout, T.; Dawson, T.L., Jr. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J. Clin. Microbiol. 2002, 40, 3350–3357.
- Gupta, A.K.; Batra, R.; Bluhm, R.; Boekhout, T.; Dawson, T.L., Jr. Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol. 2004, 51, 785–798.
- Gaitanis, G.; Velegraki, A.; Alexopoulos, E.; Chasapi, V.; Tsigonia, A.; Katsambas, A. Distribution of Malassezia species in pityriasis versicolor and seborrhoeic dermatitis in Greece. Typing of the major pityriasis versicolor isolate M. globosa. Br. J. Dermatol. 2006, 154, 854–859.
- Sugita, T.; Tajima, M.; Tsubuku, H.; Tsuboi, R.; Nishikawa, A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol. Immunol. 2006, 50, 549–552.
- Akaza, N.; Akamatsu, H.; Sasaki, Y.; Takeoka, S.; Kishi, M.; Mizutani, H.; Sano, A.; Hirokawa, K.; Nakata, S.; Matsunaga, K. Cutaneous Malassezia microbiota of healthy subjects differ by sex, body part and season. J. Dermatol. 2010, 37, 786–792.
- Clavaud, C.; Jourdain, R.; Bar-Hen, A.; Tichit, M.; Bouchier, C.; Pouradier, F.; El Rawadi, C.; Guillot, J.; Ménard-Szczebara, F.; Breton, L. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE 2013, 8, e58203.
- Tanaka, A.; Cho, O.; Saito, M.; Tsuboi, R.; Kurakado, S.; Sugita, T. Molecular characterization of the skin fungal microbiota in patients with seborrheic dermatitis. J. Clin. Exp. Dermatol. Res. 2014, 5, 239.
- Leung, M.H.; Chan, K.C.; Lee, P.K. Skin fungal community and its correlation with bacterial community of urban Chinese individuals. Microbiome 2016, 4, 1–15.
- Aydogan, K.; Tore, O.; Akcaglar, S.; Oral, B.; Ener, B.; Tunalı, S.; Saricaoglu, H. Effects of Malassezia yeasts on serum Th1 and Th2 cytokines in patients with guttate psoriasis. Int. J. Dermatol. 2013, 52, 46–52.
- Gomez-Moyano, E.; Crespo-Erchiga, V.; Martínez-Pilar, L.; Diaz, D.G.; Martínez-García, S.; Navarro, M.L.; Casaño, A.V. Do Malassezia species play a role in exacerbation of scalp psoriasis? J. Mycol. Med. 2014, 24, 87–92.
- Zhang, H.; Zhang, R.; Ran, Y.; Dai, Y.; Lu, Y.; Wang, P. Genetic polymorphism of Malassezia furfur isolates from Han and Tibetan ethnic groups in China using DNA fingerprinting. Med. Mycol. 2010, 48, 1034–1038.
- Leong, C.; Schmid, B.; Toi, M.J.; Wang, J.; Irudayaswamy, A.S.; Goh, J.P.Z.; Bosshard, P.P.; Glatz, M.; Dawson, T.L., Jr. Geographical and ethnic differences influence culturable commensal yeast diversity on healthy skin. Front. Microbiol. 2019, 10, 1891.
- Grice, E.A.; Dawson, T.L. Host–microbe interactions: Malassezia and human skin. Curr. Opin. Microbiol. 2017, 40, 81–87.
- Gaitanis, G.; Velegraki, A.; Alexopoulos, E.C.; Kapsanaki-Gotsi, E.; Zisova, L.; Ran, Y.; Zhang, H.; Arsenis, G.; Bassukas, I.D.; Faergemann, J. Malassezia furfur fingerprints as possible markers for human phylogeography. ISME J. 2009, 3, 498–502.
- Giusiano, G.; de los Angeles Sosa, M.; Rojas, F.; Vanacore, S.T.; Mangiaterra, M. Prevalence of Malassezia species in pityriasis versicolor lesions in northeast Argentina. Rev. Iberoam. Micol. 2010, 27, 71–74.
- Sugita, T.; Suzuki, M.; Goto, S.; Nishikawa, A.; Hiruma, M.; Yamazaki, T.; Makimura, K. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med. Mycol. 2010, 48, 229–233.
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64.
- Prohic, A.; Simic, D.; Sadikovic, T.J.; Krupalija-Fazlic, M. Distribution of Malassezia species on healthy human skin in Bosnia and Herzegovina: Correlation with body part, age and gender. Iran J. Microbiol. 2014, 6, 253–262.
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975.
- Oh, K.J.; Lee, S.E.; Jung, H.; Kim, G.; Romero, R.; Yoon, B.H. Detection of ureaplasmas by the polymerase chain reaction in the amniotic fluid of patients with cervical insufficiency. J. Perinat. Med. 2010, 38, 261–268.
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65.
- Dunn, A.B.; Jordan, S.; Baker, B.J.; Carlson, N.S. The maternal infant microbiome: Considerations for labor and birth. MCN. Am. J. Matern. Child Nurs. 2017, 42, 318.
- Georgountzou, A.; Papadopoulos, N.G. Postnatal innate immune development: From birth to adulthood. Front. Immunol. 2017, 8, 957.
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 1–9.
- Bazzicalupo, A.L.; Bálint, M.; Schmitt, I. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 2013, 6, 102–109.
- Gaitanis, G.; Magiatis, P.; Hantschke, M.; Bassukas, I.D.; Velegraki, A. The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 2012, 25, 106–141.
- Dupuy, A.K.; David, M.S.; Li, L.; Heider, T.N.; Peterson, J.D.; Montano, E.A.; Dongari-Bagtzoglou, A.; Diaz, P.I.; Strausbaugh, L.D. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: Discovery of Malassezia as a prominent commensal. PLoS ONE 2014, 9, e90899.
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 1–13.
- Hoggard, M.; Zoing, M.; Biswas, K.; Taylor, M.W.; Douglas, R.G. The sinonasal mycobiota in chronic rhinosinusitis and control patients. Rhinology 2019, 57, 190–199.
- Pisa, D.; Alonso, R.; Carrasco, L. Parkinson’s Disease: A Comprehensive Analysis of Fungi and Bacteria in Brain Tissue. Int. J. Biol. Sci. 2020, 16, 1135.
- Abdillah, A.; Ranque, S. Chronic Diseases Associated with Malassezia Yeast. J. Fungi 2021, 7, 855.
- Lee, K.; Zhang, I.; Kyman, S.; Kask, O.; Cope, E.K. Co-infection of Malassezia sympodialis with bacterial pathobionts Pseudomonas aeruginosa or Staphylococcus aureus leads to distinct sinonasal inflammatory responses in a murine acute sinusitis model. Front. Cell. Infect. Microbiol. 2020, 10, 472.
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358.
- Fraczek, M.G.; Chishimba, L.; Niven, R.M.; Bromley, M.; Simpson, A.; Smyth, L.; Denning, D.W.; Bowyer, P. Corticosteroid treatment is associated with increased filamentous fungal burden in allergic fungal disease. J. Allergy Clin. Immunol. 2018, 142, 407–414.
- Martinsen, E.M.; Eagan, T.M.; Leiten, E.O.; Haaland, I.; Husebø, G.R.; Knudsen, K.S.; Drengenes, C.; Sanseverino, W.; Paytuví-Gallart, A.; Nielsen, R. The pulmonary mycobiome—A study of subjects with and without chronic obstructive pulmonary disease. PLoS ONE 2021, 16, e0248967.




