Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Andres D. Klein | -- | 1817 | 2022-07-14 13:32:52 | | | |
| 2 | Beatrix Zheng | Meta information modification | 1817 | 2022-07-15 10:38:35 | | | | |
| 3 | Beatrix Zheng | -3 word(s) | 1814 | 2022-07-18 08:36:40 | | | | |
| 4 | Beatrix Zheng | -8 word(s) | 1806 | 2022-07-22 07:43:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Olguín, V.; Durán, A.; Heras, M.L.; Rubilar, J.C.; Cubillos, F.A.; Olguín, P.; Klein, A.D. Rodents as Model Organisms in Genetic Research. Encyclopedia. Available online: https://encyclopedia.pub/entry/25153 (accessed on 07 February 2026).
Olguín V, Durán A, Heras ML, Rubilar JC, Cubillos FA, Olguín P, et al. Rodents as Model Organisms in Genetic Research. Encyclopedia. Available at: https://encyclopedia.pub/entry/25153. Accessed February 07, 2026.
Olguín, Valeria, Anyelo Durán, Macarena Las Heras, Juan Carlos Rubilar, Francisco A. Cubillos, Patricio Olguín, Andrés D. Klein. "Rodents as Model Organisms in Genetic Research" Encyclopedia, https://encyclopedia.pub/entry/25153 (accessed February 07, 2026).
Olguín, V., Durán, A., Heras, M.L., Rubilar, J.C., Cubillos, F.A., Olguín, P., & Klein, A.D. (2022, July 14). Rodents as Model Organisms in Genetic Research. In Encyclopedia. https://encyclopedia.pub/entry/25153
Olguín, Valeria, et al. "Rodents as Model Organisms in Genetic Research." Encyclopedia. Web. 14 July, 2022.
Copy Citation
The advantages of using mouse models in biomedicine have been discussed extensively. Some benefits are the following: (i) the availability of genetic tools for creating disease models by transgenic, knockout, and knock-in technologies; (ii) inbred mouse strains are nearly isogenic, enabling to study how the same genetic mutation modifies a phenotype of interest in different genetic backgrounds; (iii) mouse tissues are available for omics studies which can be challenging to obtain from humans. Some limitations include different evolutive pressures for mice and humans; therefore, some systems, such as the immune system, do not function similarly in both species.
systems genetics
mouse
Drosophila
Saccharomyces cerevisiae
translational research
genetic background
precision medicine
gene mapping
1. Hybrid Mouse Diversity Panel
Currently available resources in rodents to find modifiers genes by association studies can be defined in two categories: (i) reference panels, consisting of inbred strains such as the Hybrid Mouse Diversity Panel (HMDP) and the Collaborative Cross (CC); (ii) populations derived from pseudo-random breeding of inbred strains, such as the Diversity Outbred (DO) and Heterogeneous Stock (HS) (Figure 1).
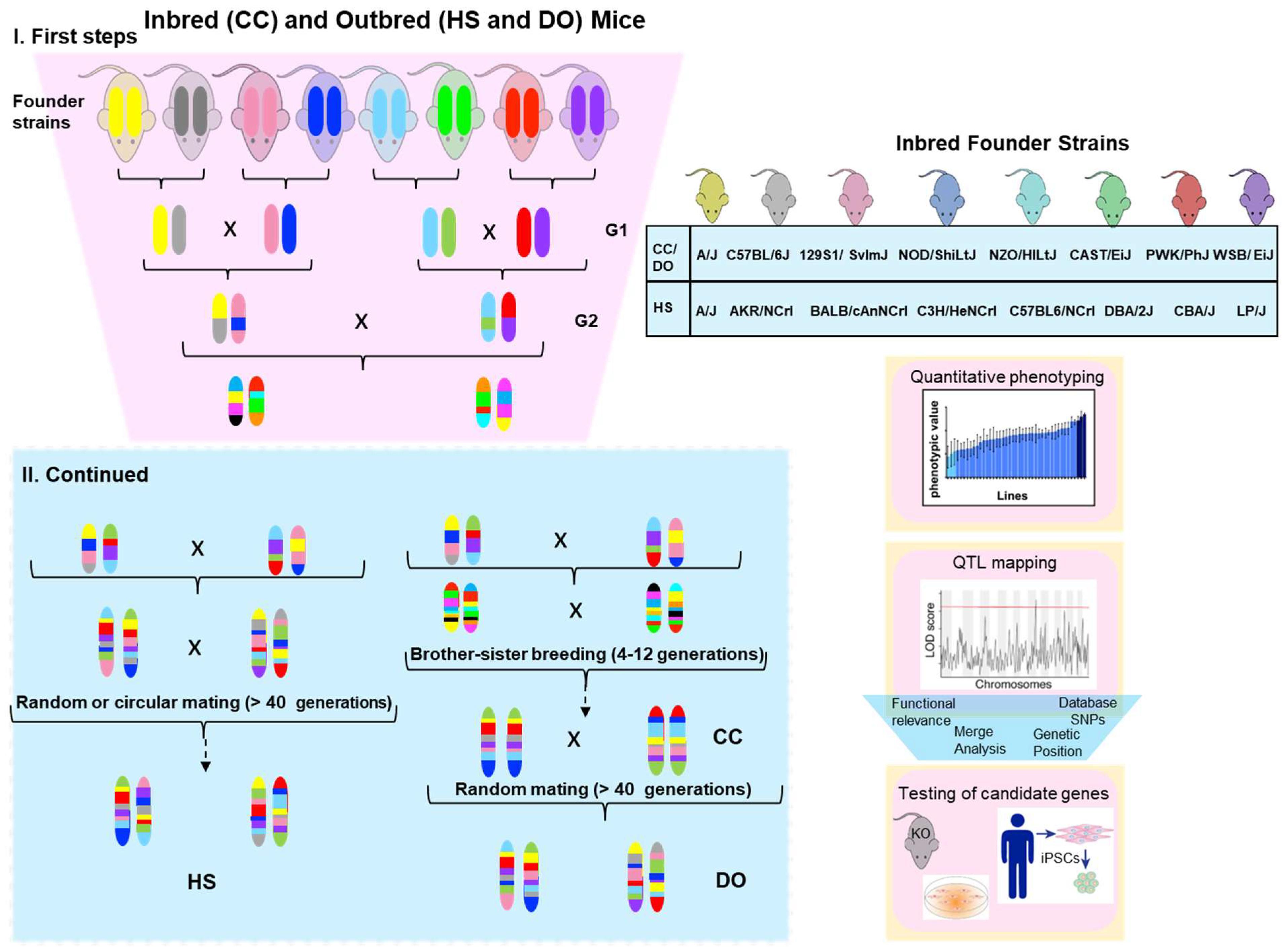
Figure 1. Breeding schemes for inbred (CC) and outbred (HS and DO) mice populations: Inbred founder strains for each panel are indicated in the right box. CC and DO populations share the same eight founder strains, five of which are standard laboratory inbred strains, while three are wild-derived strains. Colors represent the genotypes of strain chromosomes. The first steps include the combination of all eight founder genomes (outcrosses). CC is then generated as a recombinant inbred (RI) after multiple brother–sister breeding. HS and DO panels were developed as high-diversity outbred panels by over 40 generations of random outcrosses. DO was created from partially inbred Collaborative Cross (CC) mice. Quantitative phenotyping can be performed in the strains and used for gene mapping. Some signals in chromosomal locations will probably pass the threshold of significance (red line) in the LOD plot. The functional relevance of these variants can be assessed in animal models such as knockout mice and induced pluripotent stem cells (iPSC) derived from patients.
HMDP is a large panel of approximately 100 commercially available (https://www.jax.org (accessed on 22 May 2022)) and fully sequenced (www.sanger.ac.uk/science/data/mouse-genomes-project (accessed on 22 May 2022)) inbred strains: ~30 classical inbred strains and ~70 recombinant inbred (RI) strains derived mainly from crosses between C57BL/6J and DBA mice and A/J and C57BL/6J mice [1].
Advantages of using the HMDP panel are the following: (i) their genomes are known (http://mouse.cs.ucla.edu/mouseHapMap/ (accessed on 22 May 2022)); thus, it is unnecessary to spend funds performing this step; (ii) HMDP possesses ~4 million common single-nucleotide variants (SNVs), which is similar to the number present in humans [2]; (iii) high-resolution association mapping [1], which is at least an order of magnitude higher than in linkage analysis; (iv) it is possible to integrate gene mapping with other omics (transcriptomics, proteomics, and metabolomics data) [3]; (v) commercially available (from The Jackson Laboratory, Harlan, and others); (vi) sufficient bioinformatics tools for data mining of complex mouse and human disease traits, such as the Systems Genetics Resource (SGR) (http://systems.genetics.ucla.edu (accessed on 22 May 2022)); (vii) servers to perform association mapping and statistical power simulation, which are also available in R to run them in house [4].
The HMDP also has limitations. For example, extensive linkage disequilibrium (LD) blocks are observed, both within and between chromosomes, probably as a result of the selection of allelic combinations conceding higher fitness during the inbreeding [5]. Consequently, regions in LD can lead to false-positive associations in GWAS analyses. Although the HMDP has a high mapping resolution, the statistical power to detect the effect of loci is small (estimated at 50% to variants explaining 10% of the trait variance) [1]. Since most loci contributing to a complex trait have an effect size below 5% [6], variants with subtle effects cannot always be detected by the HMDP. Power can be enhanced by including additional inbred and RI strains and performing meta-analyses from other panels such as the CC or traditional crosses [7].
An exciting application of the use of mouse panels in translational research comes from crossing the classical Alzheimer’s disease (AD) mouse model (5XFAD) bearing mutations in APP and PSEN1 with 28 different strains of the BXD panel (AD-BXD). The F1 represents isogenic lines that were studied in a controlled environment. The AD-BXD panel mimicked several signs of the AD patients, including phenotypic variation in disease onset and severity. As in humans, the Apoe allele significantly affected spatial memory and other behavioral tests in the AD-BXD panel. Furthermore, hippocampal gene expression in the severe and mild lines agrees with transcriptomic changes observed in patients [8].
2. The Collaborative Cross (CC) Panel
The CC is a large panel of RI mouse strains obtained through systematically outcrossing eight founder strains, followed by randomized breeding [9]. The founder strains of the CC include five of the widely used classical inbred laboratory strains (A/J, C57BL/6J, NOD/ShiLtJ, 129S1/SvImJ, and NZO/HILtJ), as well as three wild-derived strains descendent of three M musculus subspecies (WSB, Castaneous, and PWK) (Figure 1). These eight strains have been fully sequenced and carry ~45 million SNVs, four times more than those of classical laboratory mouse strains [10].
The genomes of the CC panel are known (http://csbio.unc.edu/CCstatus/CCGenomes (accessed on 22 May 2022)), which is helpful for genetic association studies. Haplotypes can be easily visualized or reconstructed as a mosaic of the genomes of the founders [11]. Parental strains capture approximately 90% of the genetic diversity seen in the Mus musculus species [12]. This high genetic diversity significantly reduces false candidate loci. Additionally, randomized breeding substantially increases mapping resolution by reducing population structure effects [13]. CC strains have been used to map quantitative trait loci (QTLs) to less than 5 Mb intervals [14]. Online tools are available to perform GWAS and linkage analyses [15]. Several aspects of human genetics and behavioral factors can be modeled in this system, including the heterogeneities observed in neurodevelopmental disorders such as autistic spectrum disorders (ASDs) [16]. The CC panel allowed the discovery of novel candidate severity modifiers of ASD, e.g., Bai3, considered a potential target for pharmacological intervention [16].
Some considerations associated with using the CC panel are the following: (i) unique outlier phenotypes can arise in large studies, probably due to the complex genetic regulatory networks involving multiple loci with epistatic interactions [17]; in such cases, the preferred approach for identifying causal genes is traditional F2 analysis or backcrosses [18]; (ii) because identifying loci could be time-consuming, it is suggested to perform a pilot study and expand as necessary [17]; (iii) creating a panel like the CC can generate breeding complications and infertility, mainly caused by genomic incompatibility introduced by the wild-derived strains. For that reason, the initial CC project aimed to produce 1000 strains but finished with only ~100 and inspired the creation of the Diversity Outbred (DO) population.
CC lines have been used for genetic association studies of many complex traits. QTL mapping for 15 metabolism- and exercise-related traits revealed five significant loci for body weight, some of which overlapped with previous human studies [19]. Gene mapping of rotarod (exercise) performance and body weight identified 45 loci, many of them related to neurological disorders and obesity in humans, suggesting a link between physical activity and neurodegeneration [20]. A study of glucose tolerance response in the CC panel identified, only in female mice, a genomic region comprising 51 genes. This research highlighted sex differences in glucose response which should be considered in human studies [21]. The CC panel is also a valuable and reliable resource for studying host–pathogen interactions [17]. For example, to map genetic modifiers affecting the severity of Pseudomonas aeruginosa lung infections, 39 CC lines were inoculated with this pathogen. The phenotypic variability was enormous, ranging from complete resistance to lethality. It is particularly relevant to study the resistant lines since they have the biological secrets to design novel therapies for the susceptible. Genomic mapping and functional validation identified dihydropyrimidine dehydrogenase (Dpyd) and sphingosine-1-phosphate receptor 1 (S1pr1) as modifier genes. In a cohort of patients with cystic fibrosis, two SNVs in the S1PR1 gene are associated with Pseudomonas aeruginosa infection [22], again indicating the translational relevance of multigenetic background studies in animal organisms.
3. Heterogeneous Stock and Diversity Outbred Populations
Both HS and DO are high-diversity outbred mice populations. The HS was established by breeding eight inbred strains and then outbreeding them in either a circular strategy or using random crosses (Figure 1) to minimize inbreeding [23]. After 50 or more generations, the HS-generated mice were a genetic mosaic of the founders’ haplotypes [24][25]. On the other hand, the DO was established from partially inbred CC lines and is maintained indefinitely through pseudorandomized fashion non-sibling mating [26] (Figure 1). Since the DO is derived from the same eight founders as the CC, it presents the same allelic diversity as the CC strains. It can be used as a complementary tool in genetic association studies [27].
There are several advantages of using HS or DO mice compared to classical inbred mice. The outbred randomized mating increases the number of additional recombination sites compared to those of classically inbred mice; thus, each HS or DO mouse has a unique genome, which is a mosaic of the original eight founder lines, resembling human heterozygosity and allows high-resolution genetic mapping [27]. HS and DO mice have been used to finely map to intervals of 2.7 Mb [28] and less than 2 Mb [27], respectively. In addition, outbred animals are more vigorous and less prone to both early and late recessive allelic effects [29]. This genetic variability within both HS and DO populations results in a high degree of phenotypic variability; thus, outbred models enable the fine mapping of many phenotypic traits. Since the founders of CC and DO lines include wild-derived strains, unique behaviors can be observed compared to classical laboratory strains and represent a valuable tool for genetic behavior association studies [10]. A repository of DO QTL studies can be shared between laboratories (https://dodb.jax.org (accessed on 22 May 2022)). Lastly, the founders of the HS and DO populations have been sequenced [30], reducing time and expense in locating the sequences.
Alternatively, some considerations must be made in the case of HS and DO mice. Since each outbred animal is genetically and phenotypically distinct, each HS and DO mouse requires genotyping and haplotype reconstruction to perform each QTL analysis [26]. High-resolution mapping can be achieved with these panels, but analyzing many animals is necessary for sufficient statistical power, which is not always possible [31]. Candidate modifiers of wild behaviors can be identified with outbred mice. However, it is challenging to validate in these panels because each animal has a unique genotype, in contrast to inbred lines [32].
An interesting translational study using the DO panel identified a diagnostic biomarker for human tuberculosis (TB). By applying machine learning algorithms to multidimensional data, the authors discovered CXCL1 as a putative biomarker of TB in the serum of mice. The biomarker was further validated in samples derived from human patients, discriminating active TB from latent infection and non-TB lung disease [33].
References
- Bennett, B.J.; Farber, C.R.; Orozco, L.; Kang, H.M.; Ghazalpour, A.; Siemers, N.; Neubauer, M.; Neuhaus, I.; Yordanova, R.; Guan, B.; et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010, 20, 281–290.
- Lusis, A.J.; Seldin, M.M.; Allayee, H.; Bennett, B.J.; Civelek, M.; Davis, R.C.; Eskin, E.; Farber, C.; Hui, S.; Mehrabian, M.; et al. The Hybrid Mouse Diversity Panel: A resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 2016, 57, 925–942.
- Klein, A.D. Modeling diseases in multiple mouse strains for precision medicine studies. Physiol. Genom. 2017, 49, 177–179.
- Ghazalpour, A.; Rau, C.D.; Farber, C.R.; Bennett, B.J.; Orozco, L.D.; Van Nas, A.; Pan, C.; Allayee, H.; Beaven, S.W.; Civelek, M.; et al. Hybrid mouse diversity panel: A panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm. Genome 2012, 23, 680–692.
- Petkov, P.; Graber, J.; Churchill, G.A.; DiPetrillo, K.; King, B.; Paigen, K. Evidence of a Large-Scale Functional Organization of Mammalian Chromosomes. PLoS Genet. 2005, 1, e33.
- Flint, J.; Mott, R. Applying mouse complex-trait resources to behavioural genetics. Nature 2008, 456, 724–727.
- Kang, E.Y.; Han, B.; Furlotte, N.; Joo, J.W.J.; Shih, D.; Davis, R.C.; Lusis, A.J.; Eskin, E. Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice. PLoS Genet. 2014, 10, e1004022.
- Neuner, S.M.; Heuer, S.E.; Huentelman, M.J.; O’Connell, K.M.S.; Kaczorowski, C.C. Harnessing Genetic Complexity to Enhance Translatability of Alzheimer’s Disease Mouse Models: A Path toward Precision Medicine. Neuron 2019, 101, 399–411.
- Srivastava, A.; Morgan, A.P.; Najarian, M.L.; Sarsani, V.K.; Sigmon, J.S.; Shorter, J.R.; Kashfeen, A.; McMullan, R.C.; Williams, L.H.; Giusti-Rodríguez, P.; et al. Genomes of the Mouse Collaborative Cross. Genetics 2017, 206, 537–556.
- Yang, H.; Wang, J.R.; Didion, J.; Buus, R.J.; Bell, T.A.; Welsh, C.E.; Bonhomme, F.; Yu, A.H.-T.; Nachman, M.W.; Piálek, J.; et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 2011, 43, 648–655.
- Aylor, D.L.; Valdar, W.; Foulds-Mathes, W.; Buus, R.J.; Verdugo, R.A.; Baric, R.S.; Ferris, M.T.; Frelinger, J.A.; Heise, M.; Frieman, M.B.; et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011, 21, 1213–1222.
- Roberts, A.; de Villena, F.P.-M.; Wang, W.; McMillan, L.; Threadgill, D.W. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: Implications for QTL discovery and systems genetics. Mamm. Genome 2007, 18, 473–481.
- Saul, M.C.; Philip, V.M.; Reinholdt, L.G.; Chesler, E.J. High-Diversity Mouse Populations for Complex Traits. Trends Genet. 2019, 35, 501–514.
- Keele, G.; Zhang, T.; Pham, D.; Vincent, M.; Genomics, T.B.-C. Undefined Regulation of Protein Abundance in Genetically Diverse Mouse Populations; Elsevier: Amsterdam, The Netherlands, 2021.
- Ram, R.; Morahan, G. Complex Trait Analyses of the Collaborative Cross: Tools and Databases. Syst. Genet. 2016, 1488, 121–129.
- Molenhuis, R.T.; Bruining, H.; Brandt, M.J.V.; Van Soldt, P.E.; Atamni, H.J.A.-T.; Burbach, J.P.H.; Iraqi, F.A.; Mott, R.F.; Kas, M.J.H. Modeling the quantitative nature of neurodevelopmental disorders using Collaborative Cross mice. Mol. Autism 2018, 9, 63.
- Noll, K.; Ferris, M.T.; Heise, M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe 2019, 25, 484–498.
- Rogala, A.R.; Morgan, A.P.; Christensen, A.M.; Gooch, T.J.; Bell, T.A.; Miller, D.R.; Godfrey, V.L.; De Villena, F.P.-M. The Collaborative Cross as a Resource for Modeling Human Disease: CC011/Unc, a New Mouse Model for Spontaneous Colitis. Mamm. Genome 2014, 25, 95–108.
- Mathes, W.F.; Aylor, D.L.; Miller, D.R.; Churchill, G.A.; Chesler, E.J.; de Villena, F.P.-M.; Threadgill, D.W.; Pomp, D. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am. J. Physiol. Metab. 2011, 300, E1124–E1134.
- Mao, J.-H.; Langley, S.A.; Huang, Y.; Hang, M.; Bouchard, K.E.; Celniker, S.E.; Brown, J.B.; Jansson, J.; Karpen, G.H.; Snijders, A.M. Identification of genetic factors that modify motor performance and body weight using Collaborative Cross mice. Sci. Rep. 2015, 5, 16247.
- Atamni, H.J.A.-T.; Ziner, Y.; Mott, R.; Wolf, L.; Iraqi, F.A. Glucose tolerance female-specific QTL mapped in collaborative cross mice. Mamm. Genome 2016, 28, 20–30.
- Lorè, N.I.; Sipione, B.; He, G.; Strug, L.J.; Atamni, H.J.; Dorman, A.; Mott, R.; Iraqi, F.A.; Bragonzi, A. Collaborative Cross Mice Yield Genetic Modifiers for Pseudomonas aeruginosa Infection in Human Lung Disease. mBio 2020, 11, e00097–e00117.
- Woods, L.C.S. QTL mapping in outbred populations: Successes and challenges. Physiol. Genom. 2014, 46, 81–90.
- Talbot, C.J.; Nicod, A.; Cherny, S.S.; Fulker, D.W.; Collins, A.C.; Flint, J. High-resolution mapping of quantitative trait loci in outbred mice. Nat. Genet. 1999, 21, 305–308.
- Woods, L.C.S.; Mott, R. Heterogeneous Stock Populations for Analysis of Complex Traits. Syst. Genet. 2016, 1488, 31–44.
- Gatti, D.M.; Svenson, K.L.; Shabalin, A.; Wu, L.-Y.; Valdar, W.; Simecek, P.; Goodwin, N.; Cheng, R.; Pomp, D.; Palmer, A.; et al. Quantitative Trait Locus Mapping Methods for Diversity Outbred Mice. G3 Genes|Genomes|Genet. 2014, 4, 1623–1633.
- Logan, R.W.; Robledo, R.F.; Recla, J.M.; Philip, V.M.; Bubier, J.A.; Jay, J.J.; Harwood, C.; Wilcox, T.; Gatti, D.M.; Bult, C.J.; et al. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013, 12, 424–437.
- Valdar, W.; Solberg, L.C.; Gauguier, D.; Burnett, S.; Klenerman, P.; Cookson, W.O.; Taylor, M.; Rawlins, J.N.P.; Mott, R.; Flint, J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006, 38, 879–887.
- Svenson, K.L.; Gatti, D.M.; Valdar, W.; Welsh, C.E.; Cheng, R.; Chesler, E.J.; Palmer, A.A.; McMillan, L.; Churchill, G.A. High-Resolution Genetic Mapping Using the Mouse Diversity Outbred Population. Genetics 2012, 190, 437–447.
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294.
- Parker, C.C.; Palmer, A.A. Dark Matter: Are Mice the Solution to Missing Heritability? Front. Genet. 2011, 2, 32.
- Chesler, E.J. Out of the bottleneck: The Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research. Mamm. Genome 2013, 25, 3–11.
- Koyuncu, D.; Niazi, M.K.K.; Tavolara, T.; Abeijon, C.; Ginese, M.L.; Liao, Y.; Mark, C.; Specht, A.; Gower, A.C.; Restrepo, B.I.; et al. CXCL1: A new diagnostic biomarker for human tuberculosis discovered using Diversity Outbred mice. PLoS Pathog. 2021, 17, e1009773.
More
Information
Subjects:
Genetics & Heredity
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
4 times
(View History)
Update Date:
22 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




